8GTF
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C stopper-terminator complex | | Descriptor: | Head-to-tail joining protein, Major tail protein, Terminator protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTN
 
 | |
8GUF
 
 | |
8GW2
 
 | |
8GW5
 
 | |
8GWA
 
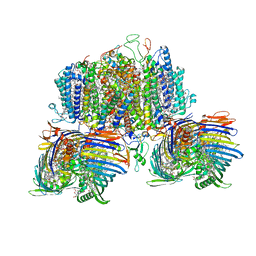 | | Structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Chen, J.H, Zhang, X. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-electron microscopy structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium.
J Integr Plant Biol, 65, 2023
|
|
8GXQ
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in MH-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X, Xu, Y. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (5.04 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
8GXS
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in H-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
8GXU
 
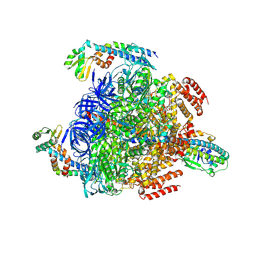 | | 1 ATP-bound V1EG of V/A-ATPase from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, SULFATE ION, V-type ATP synthase alpha chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXW
 
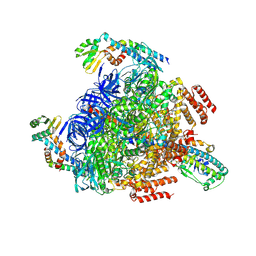 | | 2 ATP-bound V1EG of V/A-ATPase from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXX
 
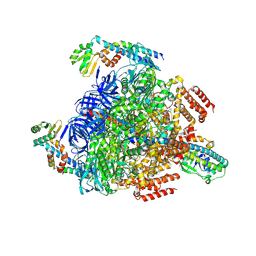 | | 3 nucleotide-bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXY
 
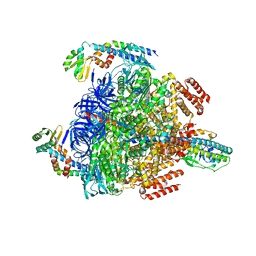 | | 2 sulfate-bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | SULFATE ION, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GXZ
 
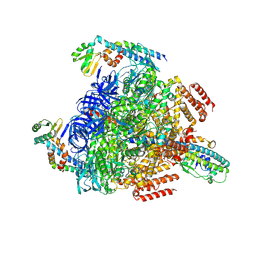 | | 1 sulfate and 1 ATP bound V1EG of V/A-ATPase from Thermus thermophilus. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Nakanishi, A, Kishikawa, J, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM analysis of V/A-ATPase intermediates reveals the transition of the ground-state structure to steady-state structures by sequential ATP binding.
J.Biol.Chem., 299, 2023
|
|
8GYM
 
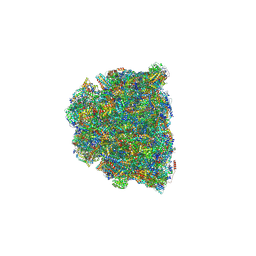 | | Cryo-EM structure of Tetrahymena thermophila respiratory mega-complex MC IV2+(I+III2+II)2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
8GZU
 
 | | Cryo-EM structure of Tetrahymena thermophila respiratory Megacomplex MC (IV2+I+III2+II)2 | | Descriptor: | 2 iron, 2 sulfur cluster-binding protein, 2-oxoglutarate/malate carrier protein, ... | | Authors: | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2022-09-27 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
8H1C
 
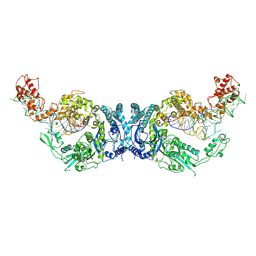 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase in complex with two tRNAs (one in tRNA binding state and the other in tRNA locked state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) (74-MER) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-10-02 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
8H1R
 
 | | Crystal structure of LptDE-YifL complex | | Descriptor: | (2R)-3-{[(2S)-3-HYDROXY-2-(PALMITOYLAMINO)PROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ... | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Lipoprotein sorting to the cell surface via a crosstalk between the Lpt and Lol pathways during outer membrane biogenesis
To Be Published
|
|
8H1S
 
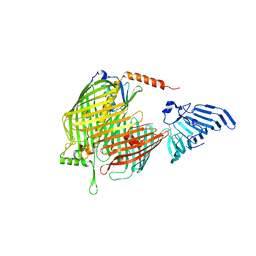 | | Crystal structure of apo-LptDE complex | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Lipoprotein sorting to the cell surface via a crosstalk between the Lpt and Lol pathways during outer membrane biogenesis
To Be Published
|
|
8H2I
 
 | | Near-atomic structure of five-fold averaged PBCV-1 capsid | | Descriptor: | MCPv1, MCPv2, MCPv3, ... | | Authors: | Shao, Q, Agarkova, I.V, Noel, E.A, Dunigan, D.D, Liu, Y, Wang, A, Guo, M, Xie, L, Zhao, X, Rossmann, M.G, Van Etten, J.L, Klose, T, Fang, Q. | | Deposit date: | 2022-10-06 | | Release date: | 2022-11-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Near-atomic, non-icosahedrally averaged structure of giant virus Paramecium bursaria chlorella virus 1.
Nat Commun, 13, 2022
|
|
8H2R
 
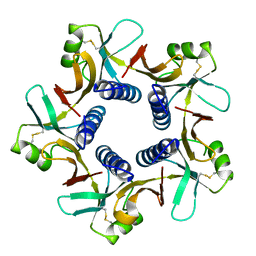 | |
8H6E
 
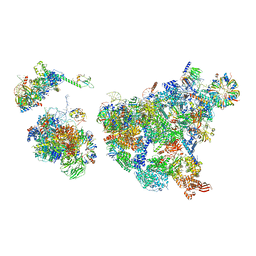 | | Cryo-EM structure of human exon-defined spliceosome in the late pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-17 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6J
 
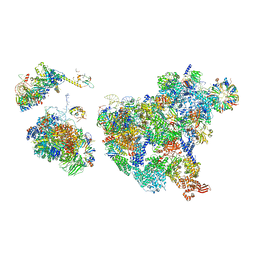 | | Cryo-EM structure of human exon-defined spliceosome in the mature pre-B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6K
 
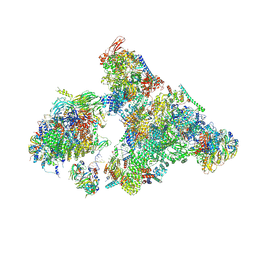 | | Cryo-EM structure of human exon-defined spliceosome in the mature B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H6L
 
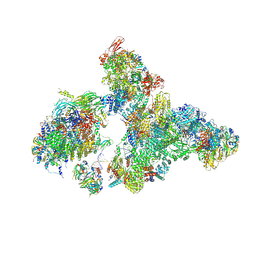 | | Cryo-EM structure of human exon-defined spliceosome in the early B state. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, W, Zhan, X, Zhang, X, Bai, R, Lei, J, Yan, C, Shi, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into human exon-defined spliceosome prior to activation.
Cell Res., 34, 2024
|
|
8H9E
 
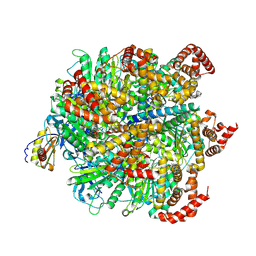 | | Human ATP synthase F1 domain, state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit O, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|






