Search
PDB entry search (PDBj Mine)
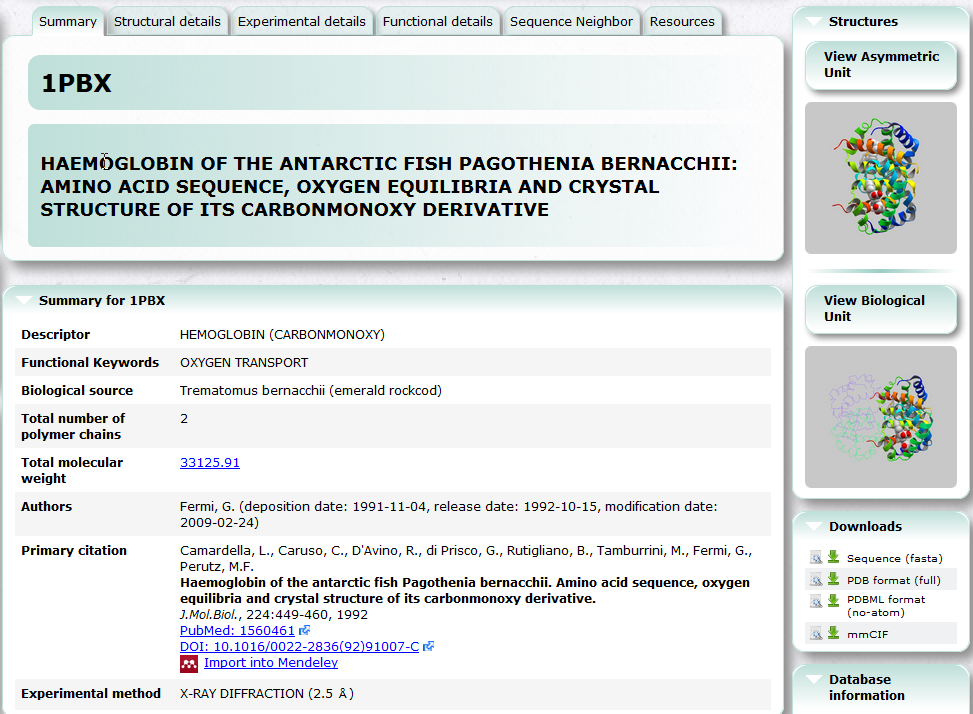
PDBj Mine is a new generic interface to search PDB entries by PDB ID and keyword etc. See also Basic Usage.
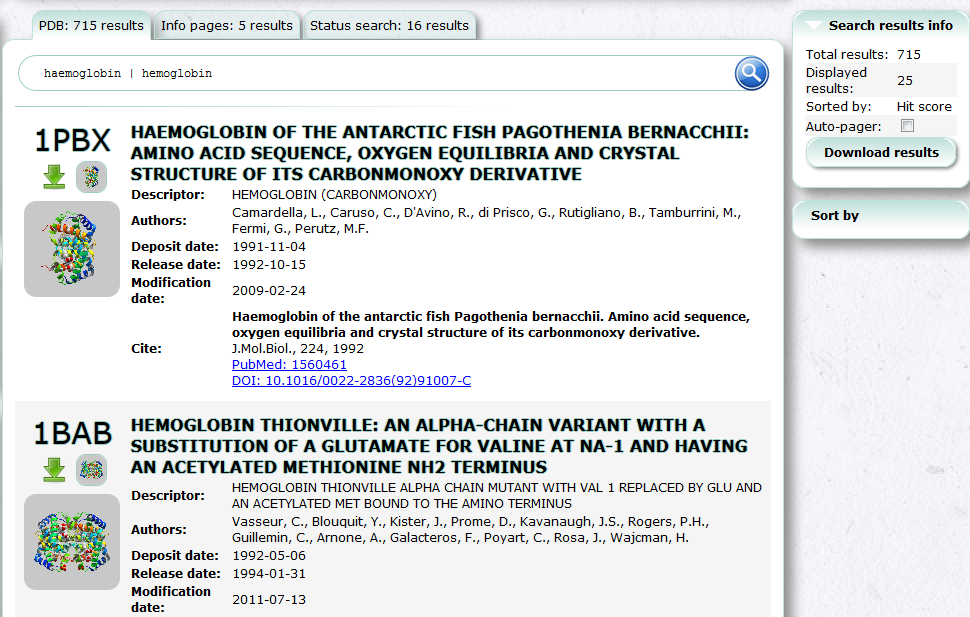
Advanced Search (PDBj Mine)
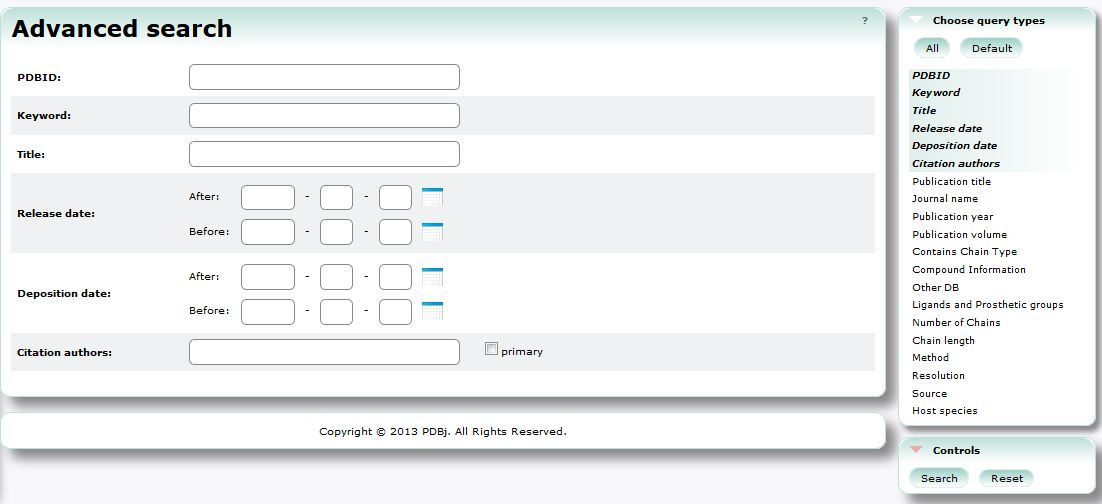
In Advanced Search, you can use combination of various conditions to search PDB entries.
Chemie search
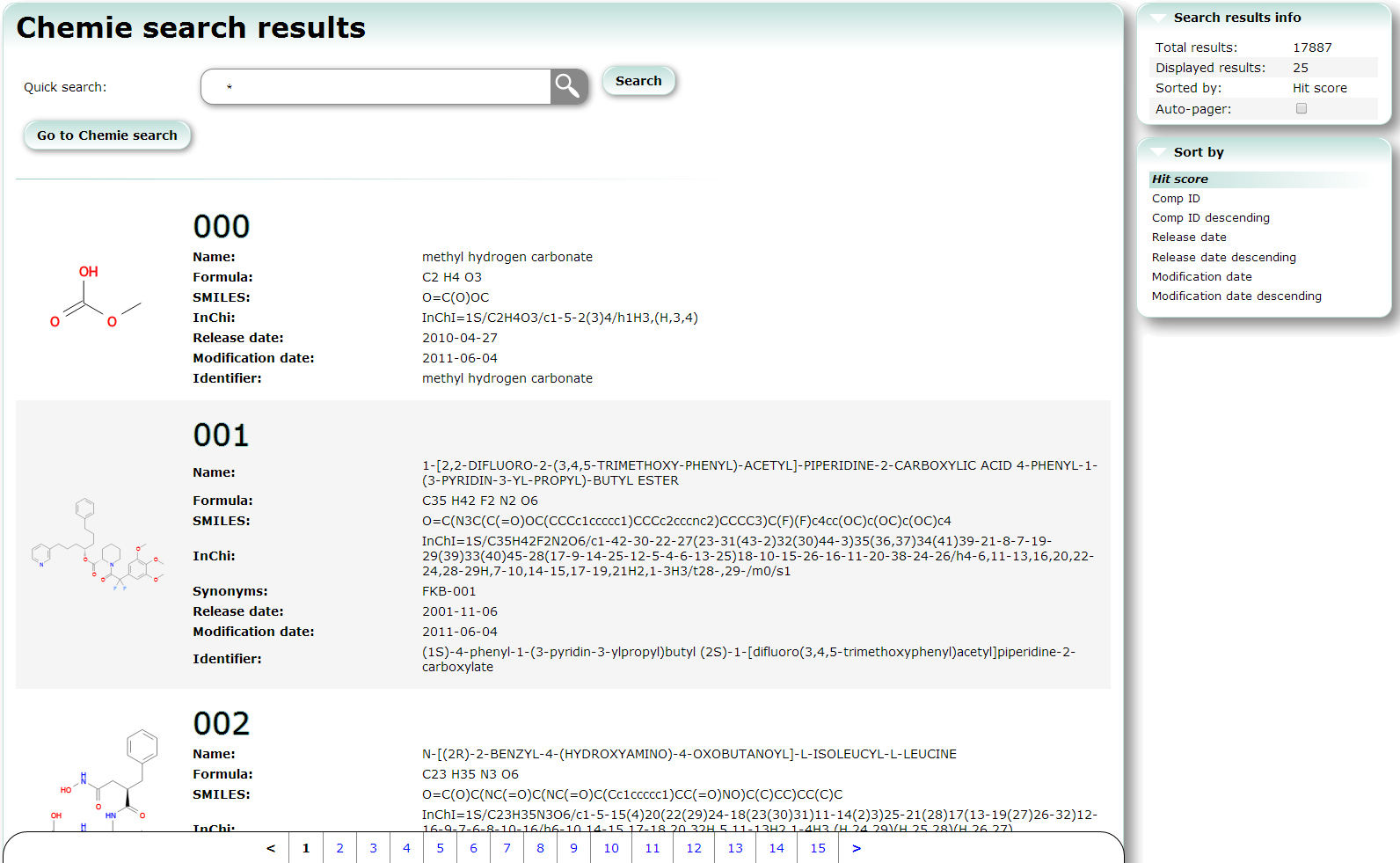
Chemie search searches non-protein and non-nucleotide compounds. Using this service, you can also find PDB entries which include the compounds.
NMR data search (BMRB)
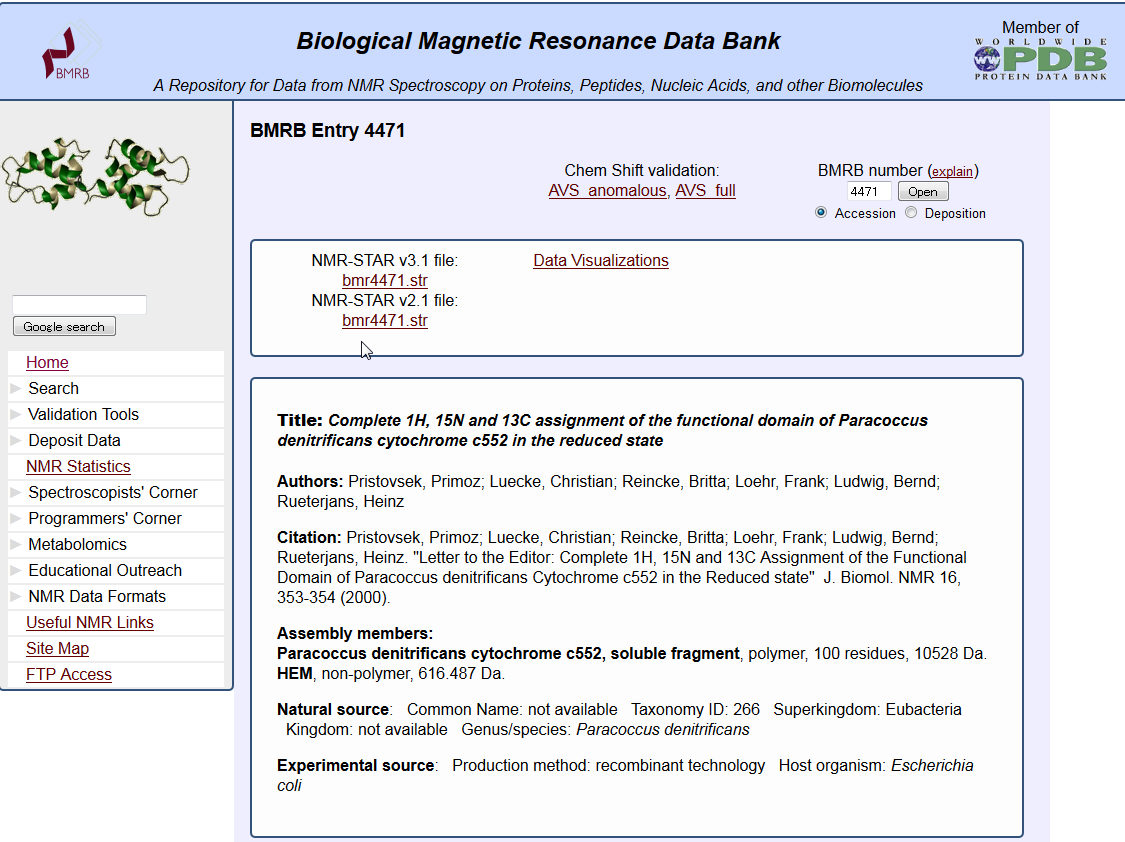
This service provides to search in the biomolecule NMR database BMRB.
Omokage Search
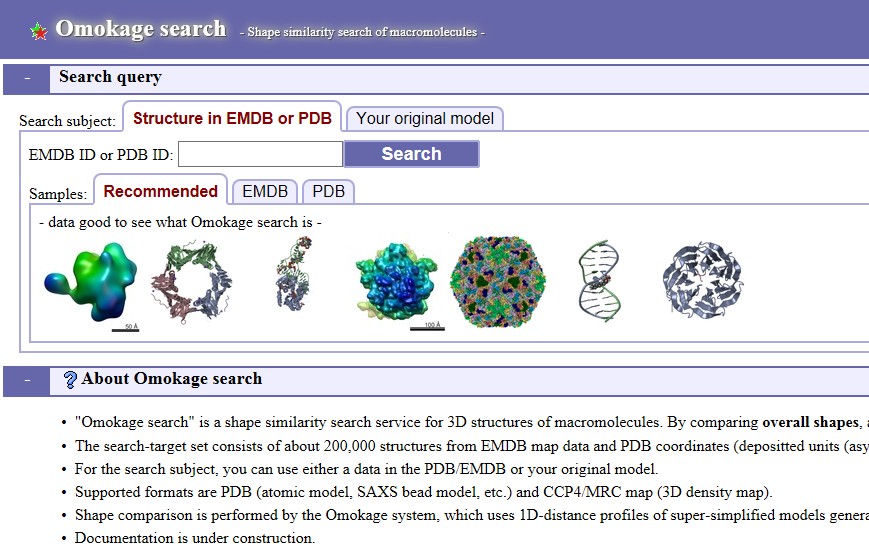
'Omokage search' is a shape similarity search service for 3D structures of macromolecules. By comparing overall shapes, and ignoring details, similar-shaped structures are searched.
See Yorodumi Docs for more details.
wwPDB/RDF
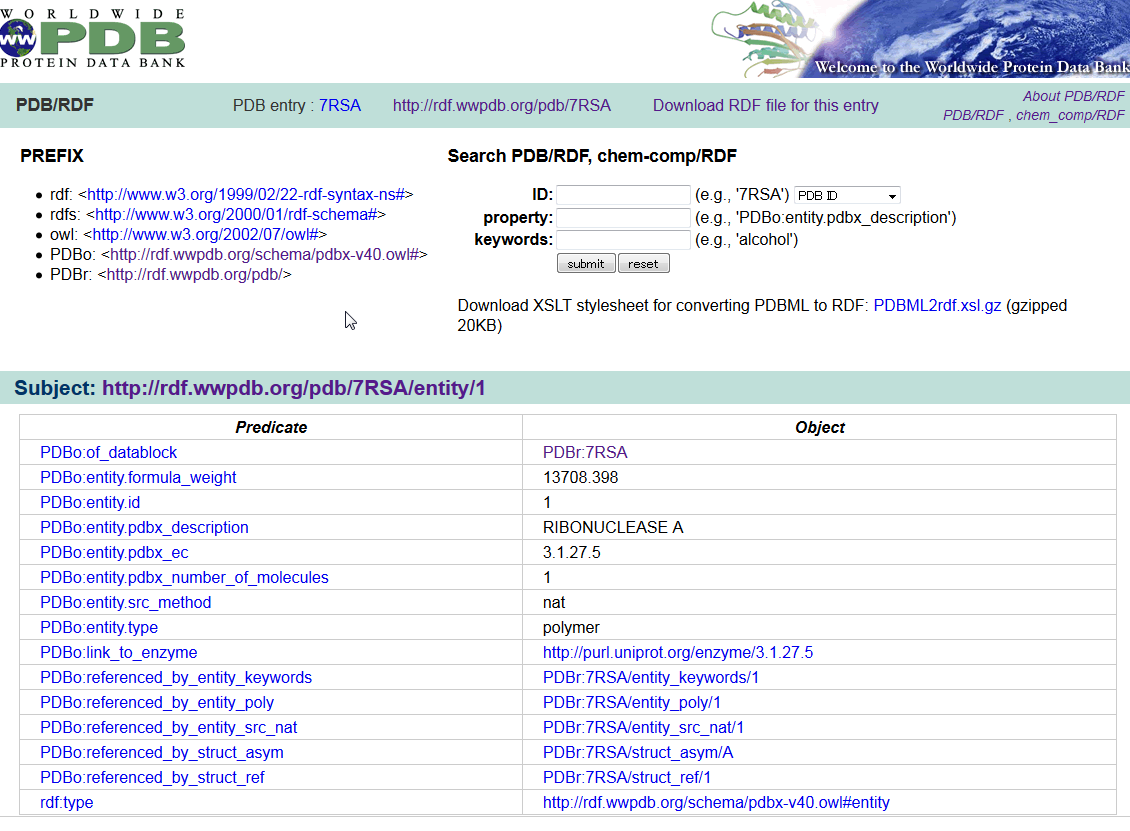
wwPDB/RDF is a collection of PDB data in the Resource Description Framework (RDF) format. The RDF format is the standard format for the Semantic Web. An ontology defined in the Web Ontology Language (OWL) is also provided for the PDB/RDF, which is a straightforward translation of the PDB mmCIF Exchange Dictionary.
URL:
http://rdf.wwpdb.org/pdb (PDB/RDF)
http://rdf.wwpdb.org/cc (chem_comp/RDF)
SeSAW
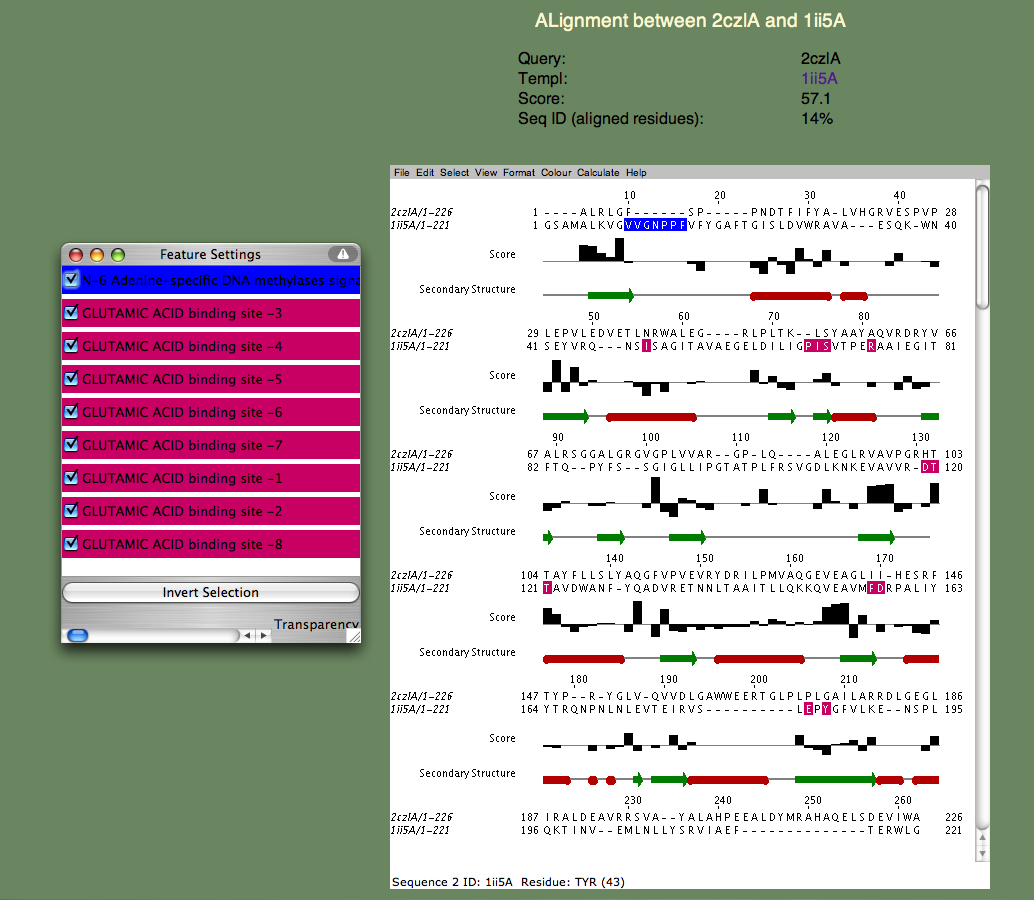
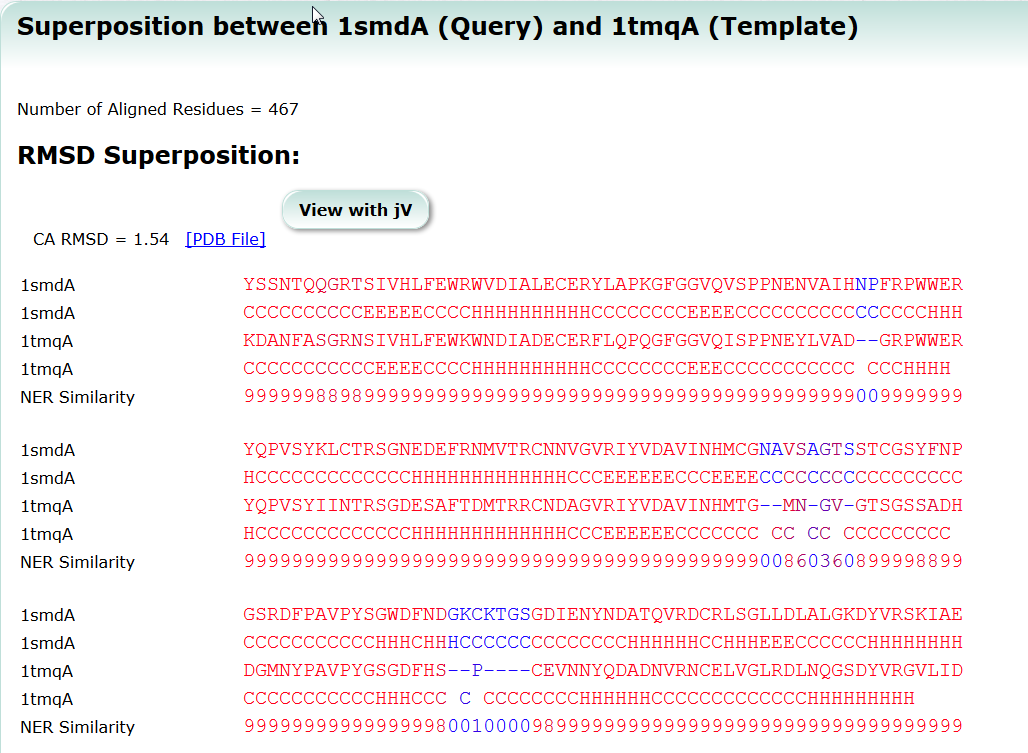
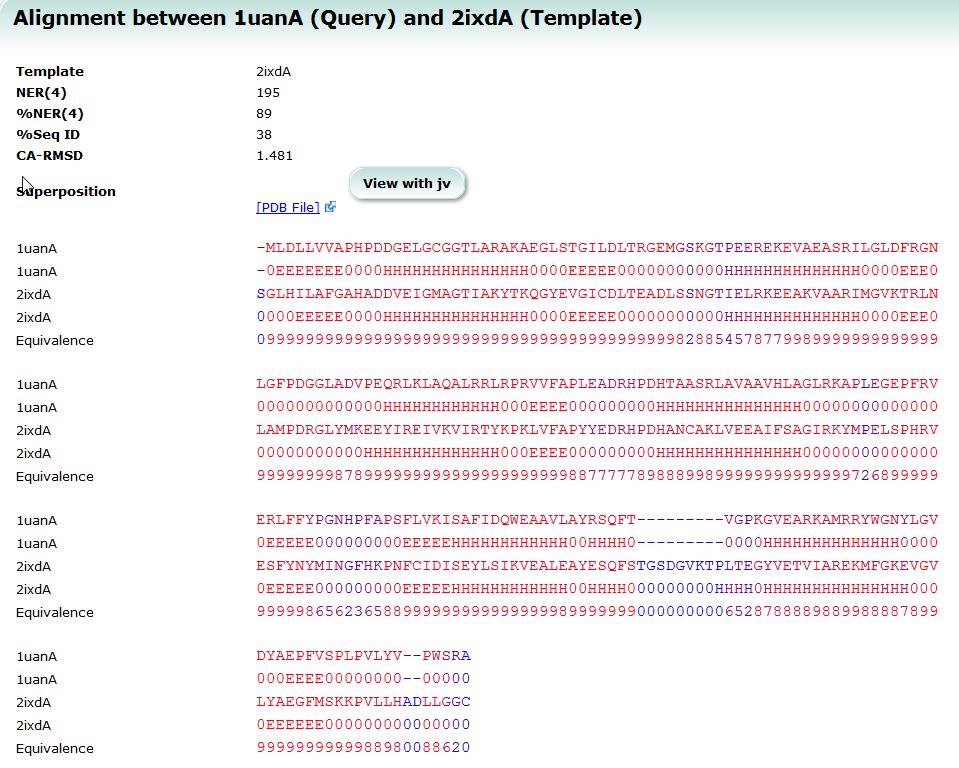
SeSAW (Sequence-Derived Structural Alignment Weights) provides functional annotations that identifies conserved sequence and structural motifs in query proteins. See also SeSAW Tutorial.
GIRAF
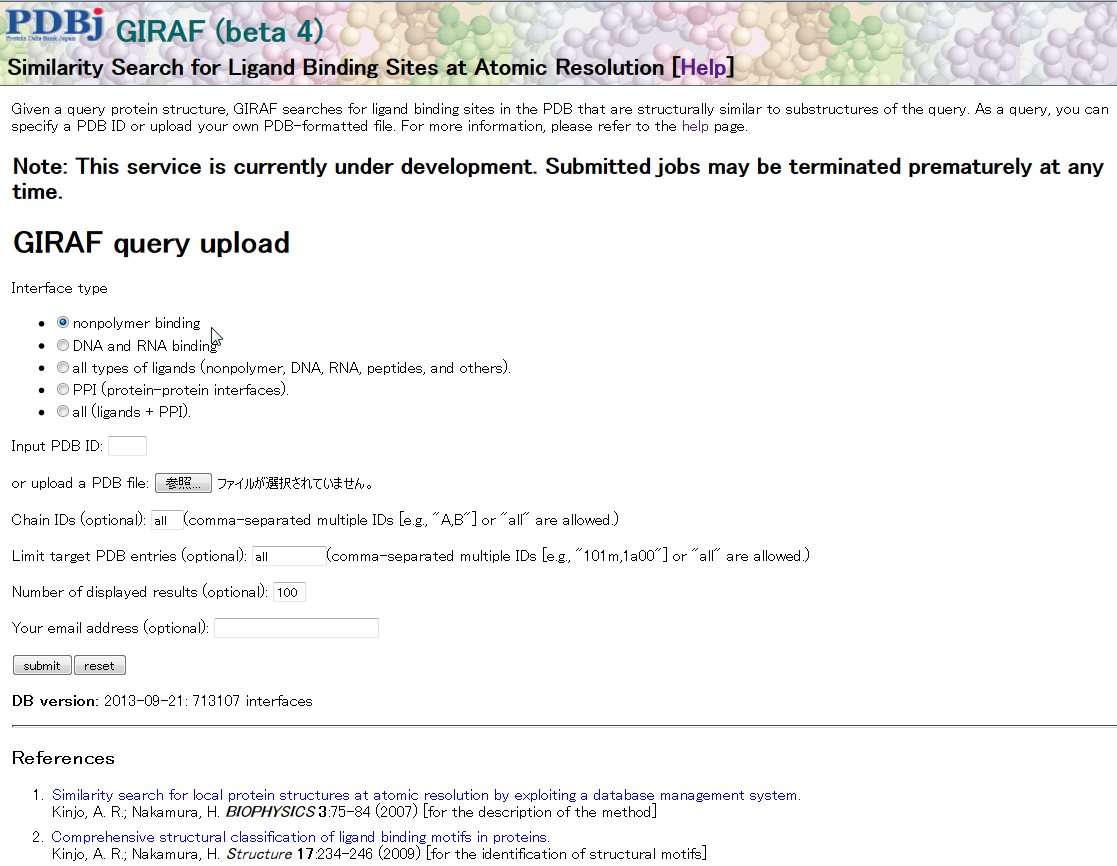
This service is currently under (active) development.
GIRAF is a similarity search service for ligand binding sites available at this site. For a given PDB ID or uploaded PDB file, GIRAF searches for known ligand binding sites in the PDB that are similar to substructures of the query.
Status Search
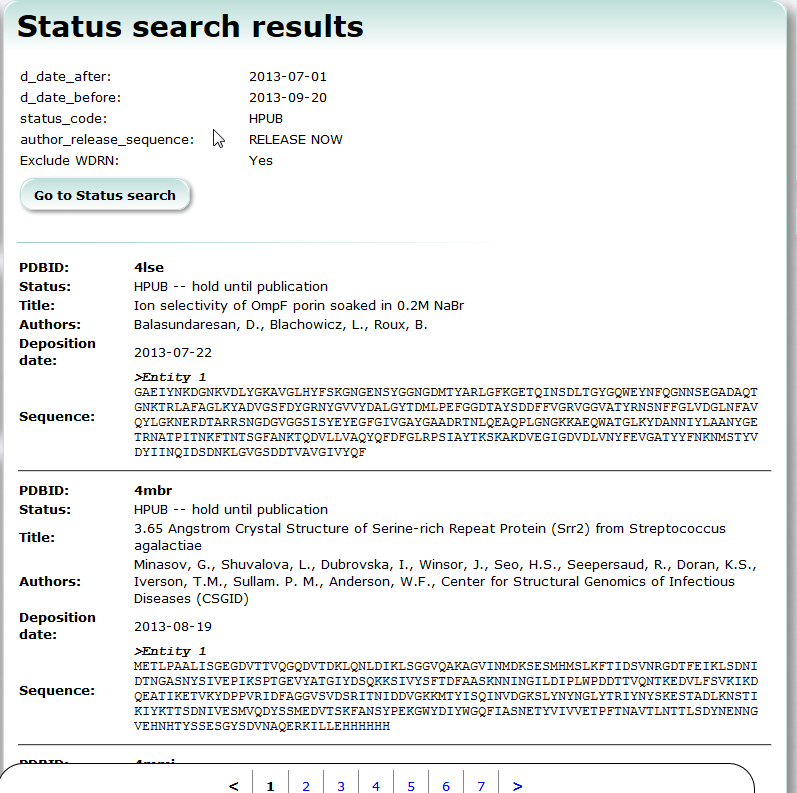
Status Search provides searching service of PDB entries which are already deposited but yet released.