9C80
 
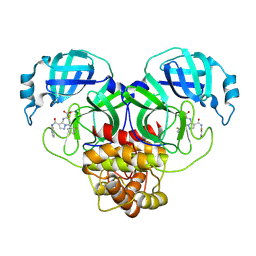 | | Co-structure of SARS-CoV-2 (COVID-19 with covalent inhibitor | | Descriptor: | (5R,7S,8R)-7-(2-fluorophenyl)-3-[(2-fluorophenyl)carbamoyl]-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-5-carboxylic acid, 3C-like proteinase nsp5 | | Authors: | Ornelas, E, Knapp, M.S. | | Deposit date: | 2024-06-11 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of Potent, Broad-Spectrum Coronavirus Main Protease Inhibitors for Pandemic Preparedness.
J.Med.Chem., 67, 2024
|
|
9C8Q
 
 | |
9CMJ
 
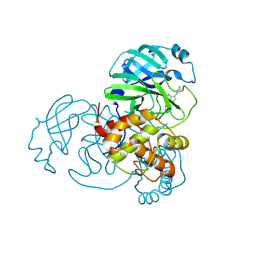
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease drug resistant mutant (L50F, E166V) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Coates, L, Gerlits, O. | | Deposit date: | 2024-07-15 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effects of SARS-CoV-2 Main Protease Mutations at Positions L50, E166, and L167 Rendering Resistance to Covalent and Noncovalent Inhibitors.
J.Med.Chem., 67, 2024
|
|
9CMN
 
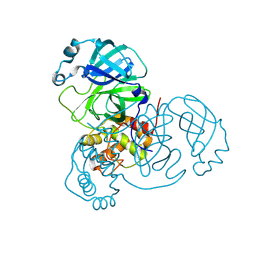 | | Room-temperature X-ray structure of SARS-CoV-2 main protease drug resistant mutant (E166A, L167F) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Coates, L, Gerlits, O. | | Deposit date: | 2024-07-15 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of SARS-CoV-2 Main Protease Mutations at Positions L50, E166, and L167 Rendering Resistance to Covalent and Noncovalent Inhibitors.
J.Med.Chem., 67, 2024
|
|
9CMS
 
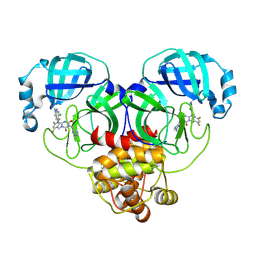 | | Room-temperature X-ray structure of SARS-CoV-2 main protease drug resistant mutant (E166V) in complex with ensitrelvir (ESV) | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Kovalevsky, A, Coates, L, Gerlits, O. | | Deposit date: | 2024-07-15 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of SARS-CoV-2 Main Protease Mutations at Positions L50, E166, and L167 Rendering Resistance to Covalent and Noncovalent Inhibitors.
J.Med.Chem., 67, 2024
|
|
9CMU
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease drug resistant mutant (L50F, E166V) in complex with ensitrelvir (ESV) | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Kovalevsky, A, Coates, L, Gerlits, O. | | Deposit date: | 2024-07-15 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of SARS-CoV-2 Main Protease Mutations at Positions L50, E166, and L167 Rendering Resistance to Covalent and Noncovalent Inhibitors.
J.Med.Chem., 67, 2024
|
|
9GS4
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54571130 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
8S6M
 
 | | SARS-CoV-2 BQ.1.1 RBD bound to the S2V29 and the S2H97 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Errico, J.M, Park, Y.J, Rietz, T, Czudnochowski, N, Nix, J.C, Cameroni, E, Corti, D, Snell, G, Marco, A.D, Pinto, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
8VIA
 
 | |
8WS3
 
 | | Crystal structure of SARS-CoV-2 Main Protease (Mpro) with covalent inhibitor 5,8-Dihydroxy-1,4-naphthoquinone | | Descriptor: | 3C-like proteinase nsp5, 5,8-bis(oxidanyl)naphthalene-1,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y, Wu, D. | | Deposit date: | 2023-10-16 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Covalent inhibitors of SARS-CoV-2 main protease
To Be Published
|
|
8WSI
 
 | |
8YF2
 
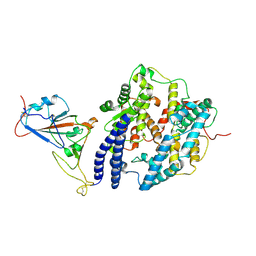 | | Cryo-EM structure of SARS-CoV-2 prototype RBD in complex with raccoon dog ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Li, L.J, Luo, C.L, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-02-23 | | Release date: | 2024-10-23 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Receptor binding and structural basis of raccoon dog ACE2 binding to SARS-CoV-2 prototype and its variants.
Plos Pathog., 20, 2024
|
|
8YFT
 
 | | Cryo-EM structure of SARS-CoV-2 alpha variant spike protein in complex with raccoon dog ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Li, L.J, Luo, C.L, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-23 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Receptor binding and structural basis of raccoon dog ACE2 binding to SARS-CoV-2 prototype and its variants.
Plos Pathog., 20, 2024
|
|
9DW6
 
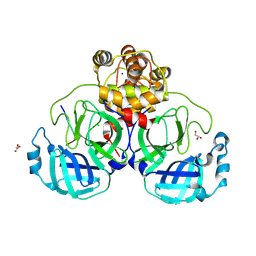 | |
8RRN
 
 | |
9GDX
 
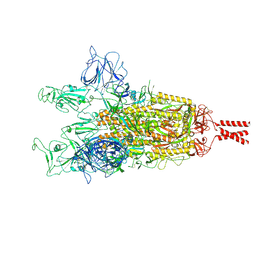 | | SARS-CoV-2 Spike protein Beta Variant at 4C structural flexibility / heterogeneity analyses | | Descriptor: | Spike glycoprotein,Fibritin | | Authors: | Herreros, D, Mata, C.P, Noddings, C, Irene, D, Agard, D.A, Tsai, M.-D, Sorzano, C.O.S, Carazo, J.M. | | Deposit date: | 2024-08-06 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Real-space heterogeneous reconstruction, refinement, and disentanglement of CryoEM conformational states with HetSIREN.
Nat Commun, 16, 2025
|
|
9GDY
 
 | | SARS-CoV-2 Spike protein Beta Variant at 37C structural flexibility / heterogeneity analyses | | Descriptor: | Spike glycoprotein,Fibritin | | Authors: | Herreros, D, Mata, C.P, Noddings, C, Irene, D, Agard, D.A, Tsai, M.-D, Sorzano, C.O.S, Carazo, J.M. | | Deposit date: | 2024-08-06 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Real-space heterogeneous reconstruction, refinement, and disentanglement of CryoEM conformational states with HetSIREN.
Nat Commun, 16, 2025
|
|
8U25
 
 | |
9CTU
 
 | | Cryo-EM structure of SARS-CoV-2 M (short conformation)bound to C1P | | Descriptor: | (2S,3R,4E)-2-(hexadecanoylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Membrane protein, POTASSIUM ION, ... | | Authors: | Dolan, K.A, Brohawn, S.G. | | Deposit date: | 2024-07-25 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Direct lipid interactions control SARS-CoV-2 M protein conformational dynamics and virus assembly.
Biorxiv, 2024
|
|
9CTW
 
 | |
9GIJ
 
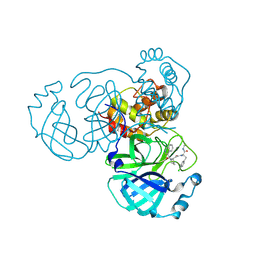 | | Crystal structure of SARS-CoV-2 Mpro with compound 5 | | Descriptor: | (2~{R})-3-(4-chlorophenyl)-2-[2-[(2~{R})-1-isoquinolin-4-ylcarbonylpyrrolidin-2-yl]ethanoyl-methyl-amino]-~{N}-methyl-propanamide, 3C-like proteinase nsp5 | | Authors: | Prasad, A, Schmitt, A, Preuss, F, Maskos, K, Wang, X, Gotchev, D, Konz Makino, D.L. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 M pro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants.
J.Med.Chem., 67, 2024
|
|
9GIL
 
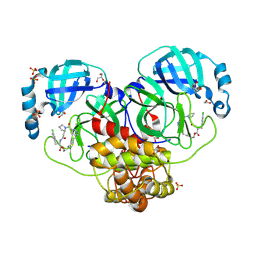 | | Crystal structure of SARS-CoV-2 Mpro with compound 12 | | Descriptor: | (7~{R},11~{R},19~{E})-11-[(4-chlorophenyl)methyl]-13-oxa-3,10,23-triazatricyclo[19.3.1.0^{3,7}]pentacosa-1(24),19,21(25),22-tetraene-2,9,12-trione, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Schmitt, A, Preuss, F, Prasad, A, Maskos, K, Wang, X, Gotchev, D, Konz Makino, D.L. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 M pro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants.
J.Med.Chem., 67, 2024
|
|
8R1Q
 
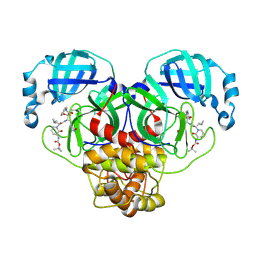 | | SARS-CoV-2 Mpro (Omicron, P132H+T169S) in complex with alpha-ketoamide 13b-K | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Sun, X, Ibrahim, M, Hilgenfeld, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-11-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8R24
 
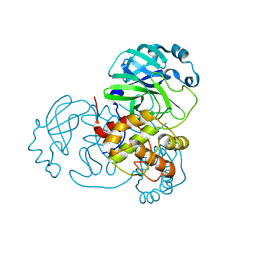 | | SARS-CoV-2 Mpro (Omicron, P132H+T169S) free enzyme | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Sun, X, Ibrahim, M, El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-11-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8R26
 
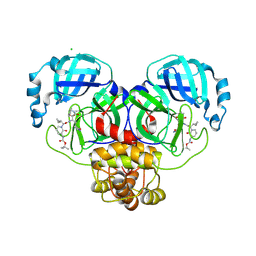 | | SARS-CoV-2 Mpro (Omicron,P132H) in complex with alpha-ketoamide 13b-K at pH 8.5 | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Sun, X, Hilgenfeld, R. | | Deposit date: | 2023-11-03 | | Release date: | 2024-11-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|








