8XCH
 
 | |
8ZPQ
 
 | |
8ZQ8
 
 | |
8ZT9
 
 | | The Crystal structure of mol066 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-propan-2-yl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]pyrimidine-2,4-dione, GLYCEROL | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2024-06-06 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of pyrimidone derivatives as nonpeptidic and noncovalent 3-chymotrypsin-like protease (3CL pro ) inhibitors with anti-coronavirus activities.
Bioorg.Chem., 154, 2025
|
|
8ZUC
 
 | | The Crystal structure of mol080 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[[6-chloranyl-2-(3-methylbutyl)indazol-5-yl]amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]pyrimidine-2,4-dione | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2024-06-08 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of pyrimidone derivatives as nonpeptidic and noncovalent 3-chymotrypsin-like protease (3CL pro ) inhibitors with anti-coronavirus activities.
Bioorg.Chem., 154, 2025
|
|
9FC2
 
 | | The crystal structure of the SARS-CoV-2 receptor binding domain in complex with the neutralizing nanobody 4. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Casasnovas, J.M, Fernandez, L.A, Silva, K. | | Deposit date: | 2024-05-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Integrating immune library probing with structure-based computational design to develop potent neutralizing nanobodies against emerging SARS-CoV-2 variants.
Mabs, 17, 2025
|
|
9GUB
 
 | | SARS-CoV-2 Mac1 in complex with MCD-628 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-(7~{H}-pyrrolo[2,3-d]pyrimidin-4-ylamino)propanoic acid, Papain-like protease nsp3 | | Authors: | Duong, M, Paakkonen, J, Lehtio, L. | | Deposit date: | 2024-09-19 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Identification of a series of pyrrolo-pyrimidine-based SARS-CoV-2 Mac1 inhibitors that repress coronavirus replication.
Mbio, 16, 2025
|
|
9NPX
 
 | |
9C7X
 
 | |
9EEI
 
 | |
9IKZ
 
 | | SARS-CoV-2 E-RTC bound to pRNA-nsp9 and GDP-BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Helicase nsp13, ... | | Authors: | Yan, L.M, Huang, Y.C, Liu, Y.X, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2024-06-29 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Substrate selection and transition reveal the mechanism for RNA capping catalyzed by SARS-CoV-2 NiRAN
To Be Published
|
|
9VAO
 
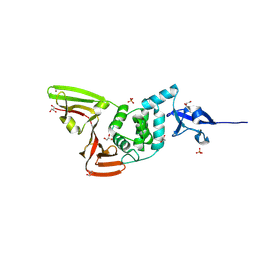 | | Crystal structure of Papain-like protease (PLpro) from SARS-CoV-2 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Papain-like protease nsp3, ... | | Authors: | Arya, R, Ganesh, J, Prashar, V, Kumar, M. | | Deposit date: | 2025-06-03 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of Papain-like protease (PLpro) from SARS-CoV-2
To Be Published
|
|
9CPP
 
 | |
9CPQ
 
 | |
9CPR
 
 | |
9CPS
 
 | |
9CPT
 
 | |
9CPU
 
 | |
9CPV
 
 | |
9CPW
 
 | |
9CPX
 
 | |
9CPY
 
 | |
9P0F
 
 | |
9CFE
 
 | |
9CFF
 
 | |








