8WFH
 
 | | Crystal structure of Omicron BA.4/5 in complex with a neutralizing antibody scFv D1 | | Descriptor: | D1 scFv, Spike protein S1 | | Authors: | Terekhov, S.S, Mokrushina, Y.A, Zhang, M, Zhang, N, Gabibov, A, Guo, Y. | | Deposit date: | 2023-09-19 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of Omicron BA.4/5 in complex with a neutralizing antibody scFv D1
To Be Published
|
|
8WFM
 
 | | Crystal structure of Omicron BA.1 in complex with a neutralizing antibody scFv T11 | | Descriptor: | Spike protein S1, T11 scFv | | Authors: | Terekhov, S.S, Mokrushina, Y.A, Zhang, M, Zhang, N, Gabibov, A, Guo, Y. | | Deposit date: | 2023-09-19 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of Omicron BA.1 in complex with a neutralizing antibody scFv T11
To Be Published
|
|
9DN4
 
 | | Crystal structure of a SARS-CoV-2 20-mer RNA in complex with FAB BL3-6S97N | | Descriptor: | CHLORIDE ION, FAB BL3-6S97N HEAVY CHAIN, FAB BL3-6S97N LIGHT CHAIN, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Hegde, S, Wang, J. | | Deposit date: | 2024-09-16 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic studies of small molecule ligands selective to RNA single G bulges.
Nucleic Acids Res., 53, 2025
|
|
8WKE
 
 | |
8WSJ
 
 | |
8WSK
 
 | |
8K18
 
 | |
8K19
 
 | |
8WXL
 
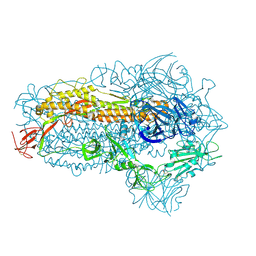 | | Structure of the SARS-CoV-2 BA.2.86 spike glycoprotein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-10-30 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XUX
 
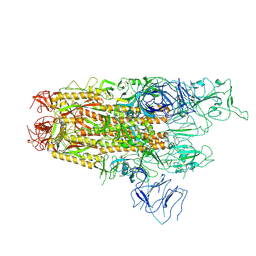 | | Structure of the SARS-CoV-2 BA.2.86 spike protein (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XV0
 
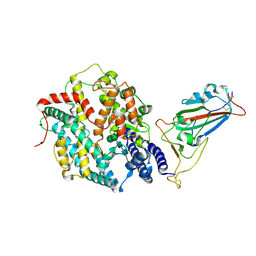 | | Structure of SARS-CoV-2 BA.2.86 spike RBD in complex with ACE2 (up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XVM
 
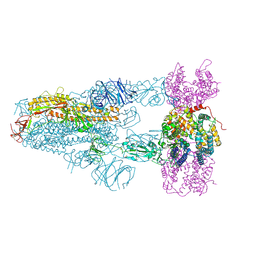 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (3-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
9IU1
 
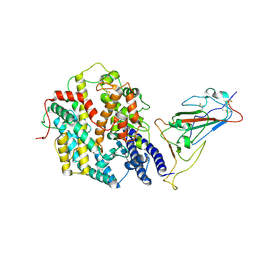 | | Structure of SARS-CoV-2 JN.1 spike RBD in complex with ACE2 (up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-07-20 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8WPW
 
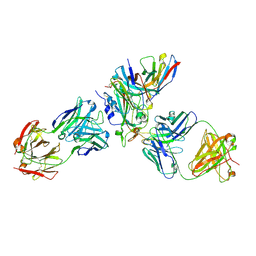 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 receptor-binding domain (RBD) complexed with CB6 mutant,S309, and S304 antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 fab mutant heavy chain, CB6 fab mutant light chain, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Enabling the immune escaped etesevimab fully-armed against SARS-CoV-2 Omicron subvariants including KP.2
Hlife, 3, 2025
|
|
8WPY
 
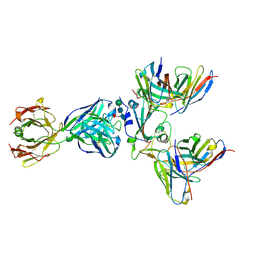 | | Cryo-EM structure of SARS-CoV-2 receptor-binding domain (RBD) complexed with CB6 mutant,S309, and S304 antibodies | | Descriptor: | CB6 fab mutant heavy chain, CB6 fab mutant light chain, S304 fab heavy chain, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Enabling the immune escaped etesevimab fully-armed against SARS-CoV-2 Omicron subvariants including KP.2
Hlife, 3, 2025
|
|
9ATM
 
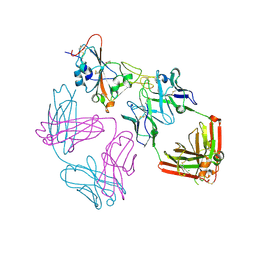 | | SARS-CoV-2 EG.5 RBD bound to the VIR-7229 and the S2H97 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rietz, T, Park, Y.J, Errico, J, Czudnochowski, N, Nix, J.C, Corti, D, Snell, G, Marco, A.D, Pinto, D, Cameroni, E, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-27 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
9AU1
 
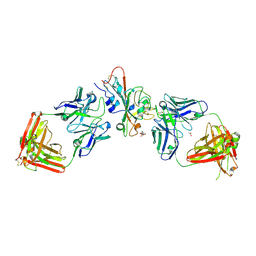 | | SARS-CoV-2 XBB.1.5 RBD bound to the VIR-7229 and the S309 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Rietz, T, Park, Y.J, Errico, J, Czudnochowski, N, Nix, J.C, Corti, D, Snell, G, Marco, A.D, Pinto, D, Cameroni, E, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-27 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
9AU2
 
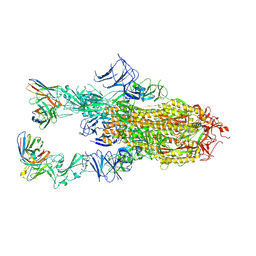 | | VIR-7229 Fab fragment bound the BA.2.86 spike trimer (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Park, Y.J, Veelser, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
9CMN
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease drug resistant mutant (E166A, L167F) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Kovalevsky, A, Coates, L, Gerlits, O. | | Deposit date: | 2024-07-15 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of SARS-CoV-2 Main Protease Mutations at Positions L50, E166, and L167 Rendering Resistance to Covalent and Noncovalent Inhibitors.
J.Med.Chem., 67, 2024
|
|
9GS4
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54571130 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
8S6M
 
 | | SARS-CoV-2 BQ.1.1 RBD bound to the S2V29 and the S2H97 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Errico, J.M, Park, Y.J, Rietz, T, Czudnochowski, N, Nix, J.C, Cameroni, E, Corti, D, Snell, G, Marco, A.D, Pinto, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
8VIA
 
 | |
8YF2
 
 | | Cryo-EM structure of SARS-CoV-2 prototype RBD in complex with raccoon dog ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Li, L.J, Luo, C.L, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-02-23 | | Release date: | 2024-10-23 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Receptor binding and structural basis of raccoon dog ACE2 binding to SARS-CoV-2 prototype and its variants.
Plos Pathog., 20, 2024
|
|
8YFT
 
 | | Cryo-EM structure of SARS-CoV-2 alpha variant spike protein in complex with raccoon dog ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Li, L.J, Luo, C.L, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-23 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Receptor binding and structural basis of raccoon dog ACE2 binding to SARS-CoV-2 prototype and its variants.
Plos Pathog., 20, 2024
|
|
9DW6
 
 | |








