8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8SGU
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-13 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8SMI
 
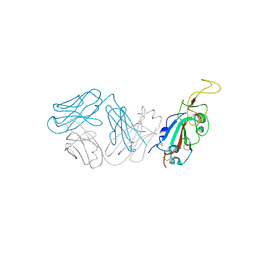 | | Crystal structure of antibody WRAIR-2123 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2123 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8WOX
 
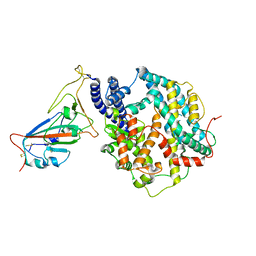 | | Cryo-EM structure of SARS-CoV-2 prototype RBD in complex with rabbit ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | Deposit date: | 2023-10-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8WOY
 
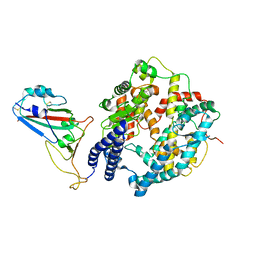 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with rabbit ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, L.J, Shi, K.Y, Yu, G.H, Gao, G.F. | | Deposit date: | 2023-10-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of increased binding affinities of spikes from SARS-CoV-2 Omicron variants to rabbit and hare ACE2s reveals the expanding host tendency.
Mbio, 15, 2024
|
|
8WTD
 
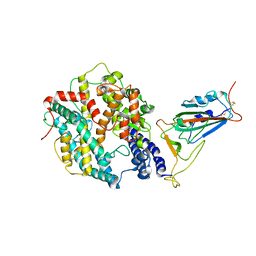 | | XBB.1.5.10 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTJ
 
 | | XBB.1.5.70 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.64 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8HPF
 
 | |
8HPU
 
 | |
8HQ7
 
 | |
8HQI
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8HQJ
 
 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8HQT
 
 | | The complex structure of COPI cargo sorting module with SARS-CoV-2 Spike KxHxx sorting motif | | Descriptor: | Coatomer subunit beta', SARS-CoV-2 Spike KxHxx motif | | Authors: | Ma, W.F, Nan, Y.N, Yang, M.R, Li, Y.Q. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 spike host cell surface exposure promoted by a COPI sorting inhibitor.
Acta Pharm Sin B, 13, 2023
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HRI
 
 | |
8HRL
 
 | | SARS-CoV-2 Delta S-RBD-ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Xu, J, Meng, F, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Self-assembled superstructure alleviates air-water interface effect in cryo-EM.
Nat Commun, 15, 2024
|
|
8HSM
 
 | | CRYSTAL STRUCTURE OF BAT MHC CLASS I MYLU-B-67 | | Descriptor: | Beta-2-microglobulin, Ig-like domain-containing protein, PHE-PRO-GLN-SER-ALA-PRO-HIS-GLY-VAL | | Authors: | Wang, S.Q, Zhang, N.Z. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Amino acid insertion in Bat MHC-I enhances complex stability and augments peptide presentation.
Commun Biol, 7, 2024
|
|
8HSO
 
 | |
8HSW
 
 | | CRYSTAL STRUCTURE OF BAT MHC CLASS I MYLU-B-67 FOR 2.1 ANGSTROM | | Descriptor: | Beta-2-microglobulin, Ig-like domain-containing protein, THR-PRO-GLN-SER-ALA-PRO-HIS-GLY-VAL | | Authors: | Wang, S.Q, Zhang, N.Z. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Amino acid insertion in Bat MHC-I enhances complex stability and augments peptide presentation.
Commun Biol, 7, 2024
|
|
8HT1
 
 | |
8HT9
 
 | | CRYSTAL STRUCTURE OF BAT MHC CLASS I MYLU-B-67 FOR 2.2 ANGSTROM | | Descriptor: | Beta-2-microglobulin, GLU-PRO-GLN-SER-ALA-PRO-HIS-GLY-VAL, Ig-like domain-containing protein | | Authors: | Wang, S.Q, Zhang, N.Z. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Amino acid insertion in Bat MHC-I enhances complex stability and augments peptide presentation.
Commun Biol, 7, 2024
|
|
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q93
 
 | | Crystal structure of the SARS-COV-2 RBD with neutralizing-VHHs Re30H02 and Re21D01 | | Descriptor: | Nanobody Re21D01, Nanobody Re30H02, Spike protein S1 | | Authors: | Aksu, M, Guttler, T, Rymarenko, O, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q94
 
 | |
8Q95
 
 | | Crystal structure of the SARS-CoV-2 BA.1 RBD with neutralizing-VHHs Ma16B06 and Ma3F05 | | Descriptor: | Nanobody Ma16B06, Nanobody Ma3F05, Spike protein S1 | | Authors: | Aksu, M, Rymarenko, O, Guttler, T, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|








