7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7R7N
 
 | |
7M42
 
 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of neutralizing antibodies REGN10985 and REGN10989 | | Descriptor: | REGN10985 antibody Fab fragment heavy chain, REGN10985 antibody Fab fragment light chain, REGN10989 antibody Fab fragment heavy chain, ... | | Authors: | Zhou, Y, Romero Hernandez, A, Saotome, K, Franklin, M.C. | | Deposit date: | 2021-03-19 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies.
Cell, 184, 2021
|
|
7MLZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | Descriptor: | B1-182.1 Fab heavy chain, B1-182.1 Fab light chain, Spike protein S1, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
7MM0
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with neutralizing antibody B1-182.1 that targets the receptor-binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B1-182.1 Fab heavy chain, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N0B
 
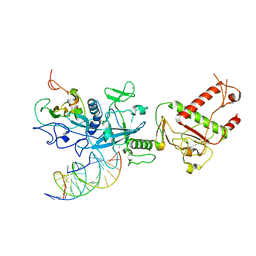 | | Cryo-EM structure of SARS-CoV-2 nsp10-nsp14 (WT)-RNA complex | | Descriptor: | CALCIUM ION, Non-structural protein 10, Proofreading exoribonuclease, ... | | Authors: | Liu, C, Yang, Y. | | Deposit date: | 2021-05-25 | | Release date: | 2021-07-28 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme.
Science, 373, 2021
|
|
7N0C
 
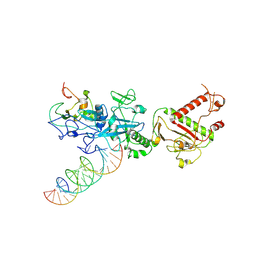 | |
7N0D
 
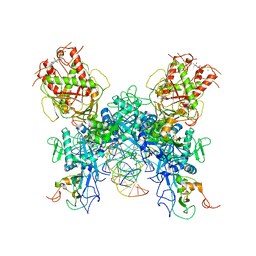 | |
7N1A
 
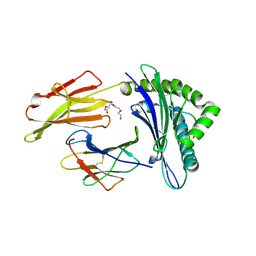 | | SARS-CoV-2 YLQ peptide binds to HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1B
 
 | | SARS-CoV-2 RLQ peptide binds to HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1E
 
 | | SARS-CoV-2 RLQ peptide-specific TCR pRLQ3 binds to RLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1F
 
 | | SARS-CoV-2 YLQ peptide-specific TCR pYLQ7 binds to YLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N6D
 
 | | HLA peptide complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Chaurasia, P, Petersen, J, Rossjohn, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of biased T cell receptor recognition of an immunodominant HLA-A2 epitope of the SARS-CoV-2 spike protein.
J.Biol.Chem., 297, 2021
|
|
7N6E
 
 | | TCR peptide HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Spike protein S1, ... | | Authors: | Chaurasia, P, Rossjohn, J, Petersen, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of biased T cell receptor recognition of an immunodominant HLA-A2 epitope of the SARS-CoV-2 spike protein.
J.Biol.Chem., 297, 2021
|
|
7P3D
 
 | | MHC I A02 Allele presenting YLQPRTFLL | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Emergence of immune escape at dominant SARS-CoV-2 killer T cell epitope.
Cell, 185, 2022
|
|
7F62
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25 (Focused refinement of S-RBD and chAb-25 region) | | Descriptor: | RBD-chAb-25, Heavy chain, Light chain, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-06-24 | | Release date: | 2021-08-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-guided antibody cocktail for prevention and treatment of COVID-19.
Plos Pathog., 17, 2021
|
|
7KM5
 
 | | Crystal structure of SARS-CoV-2 RBD complexed with Nanosota-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Spike protein S1, ... | | Authors: | Ye, G, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The development of Nanosota - 1 as anti-SARS-CoV-2 nanobody drug candidates.
Elife, 10, 2021
|
|
7N3I
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment C098 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C098 Fab heavy chain, C098 Fab light chain, ... | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N62
 
 | | SARS-CoV-2 Spike (2P) in complex with C12C9 Fab (NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C12C9 Fab heavy chain, C12C9 Fab light chain, ... | | Authors: | Windsor, I.W, Jenni, S, Bajic, G, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
7N64
 
 | | SARS-CoV-2 Spike (2P) in complex with G32R7 Fab (RBD and NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G32R7 Fab heavy chain, ... | | Authors: | Windsor, I.W, Jenni, S, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
7N9A
 
 | |
7N9B
 
 | |
7N9C
 
 | |
7N9E
 
 | |
7P77
 
 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 3up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|








