7K9J
 
 | |
7K9K
 
 | |
7RBR
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with a Lys48-linked di-ubiquitin | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Papain-like protease, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-06 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7RBS
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with human ISG15 | | 分子名称: | Papain-like protease, Ubiquitin-like protein ISG15, ZINC ION | | 著者 | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-06 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7RBZ
 
 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-017-20 | | 分子名称: | 3C-like proteinase, 5-chloropyridin-3-yl 2,3-dihydro-1H-indole-4-carboxylate | | 著者 | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-06 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
7RC0
 
 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-091-20 | | 分子名称: | 3C-like proteinase, 5-chloro-4-methylpyridin-3-yl 1H-indole-4-carboxylate, SODIUM ION | | 著者 | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-06 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
7MZF
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 37 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | 著者 | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.493 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZG
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 42 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PDI 42 heavy chain, ... | | 著者 | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZH
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 119 | | 分子名称: | Spike protein S1, WCSL 119 heavy chain, WCSL 119 light chain, ... | | 著者 | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZI
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 | | 分子名称: | GLYCEROL, Spike protein S1, TETRAETHYLENE GLYCOL, ... | | 著者 | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZJ
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 and Fab PDI 93 | | 分子名称: | GLYCEROL, PDI 93 heavy chain, PDI 93 light chain, ... | | 著者 | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZK
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 and Fab PDI 96 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | 著者 | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZL
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 210 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 210 heavy chain, PDI 210 light chain, ... | | 著者 | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZM
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 215 | | 分子名称: | ISOPROPYL ALCOHOL, PDI 215 heavy chain, PDI 215 light chain, ... | | 著者 | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZN
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 231 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 231 heavy chain, PDI 231 light chain, ... | | 著者 | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7N4I
 
 | |
7N4J
 
 | |
7NTS
 
 | | Crystal structure of the SARS-CoV-2 Main Protease with oxidized C145 | | 分子名称: | DIMETHYL SULFOXIDE, FORMIC ACID, GLYCEROL, ... | | 著者 | Dupre, E, Villeret, V, Hanoulle, X. | | 登録日 | 2021-03-10 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.477 Å) | | 主引用文献 | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7P51
 
 | |
7RR0
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 222 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 222 Fab Heavy Chain, PDI 222 Fab Light Chain, ... | | 著者 | Pymm, P, Glukhova, A, Black, K.A, Tham, W.H. | | 登録日 | 2021-08-08 | | 公開日 | 2021-10-06 | | 最終更新日 | 2021-10-27 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7RXD
 
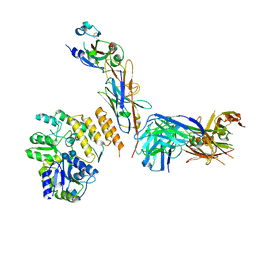 | | CryoEM structure of RBD domain of COVID-19 in complex with Legobody | | 分子名称: | Fab_8D3_2 heavy chain, Fab_8D3_2 light chain, Maltodextrin-binding protein,Immunoglobulin G-binding protein A,Immunoglobulin G-binding protein G, ... | | 著者 | Wu, X.D, Rapoport, T.A. | | 登録日 | 2021-08-22 | | 公開日 | 2021-10-06 | | 最終更新日 | 2021-10-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure determination of small proteins by nanobody-binding scaffolds (Legobodies).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7S0B
 
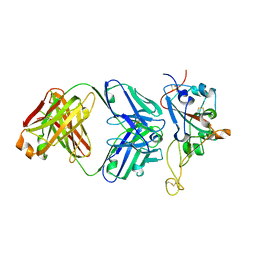 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody N-612-056 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-612-056 Fab Heavy Chain, N-612-056 Light Chain, ... | | 著者 | Tanaka, S, Barnes, C.O, Bjorkman, P.J. | | 登録日 | 2021-08-30 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Rapid identification of neutralizing antibodies against SARS-CoV-2 variants by mRNA display.
Cell Rep, 38, 2022
|
|
7S0C
 
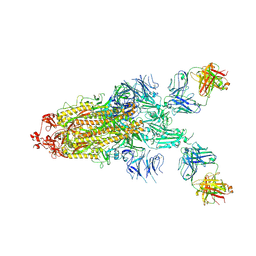 | |
7S0D
 
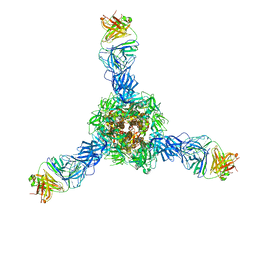 | |
7S0E
 
 | |







