7NX6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab Heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX7
 
 | | Crystal structure of the K417N mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX8
 
 | | Crystal structure of the K417T mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX9
 
 | | Crystal structure of the N501Y mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 B.1.351 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXB
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 P.1 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXC
 
 | |
7L56
 
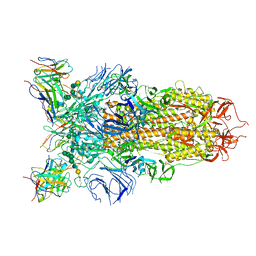 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-43 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2-43 variable domain heavy chain, ... | | 著者 | Rapp, M, Shapiro, L. | | 登録日 | 2020-12-21 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7L57
 
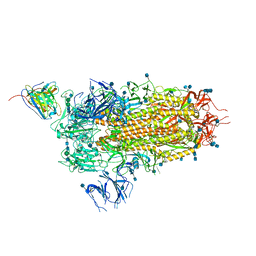 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-15 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Rapp, M, Shapiro, L. | | 登録日 | 2020-12-21 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (5.87 Å) | | 主引用文献 | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7L58
 
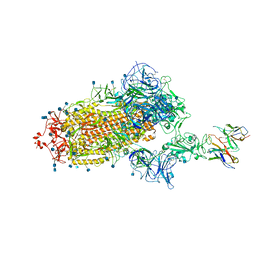 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab H4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab H4 variable domain heavy chain, ... | | 著者 | Rapp, M, Shapiro, L. | | 登録日 | 2020-12-21 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (5.07 Å) | | 主引用文献 | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7LKD
 
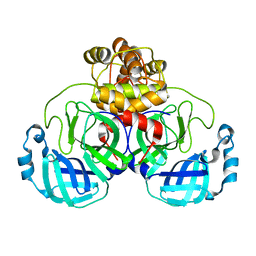 | |
7LKE
 
 | |
7LMC
 
 | |
7LXW
 
 | |
7LXX
 
 | |
7LXY
 
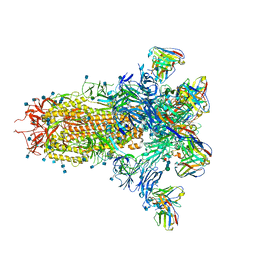 | |
7LXZ
 
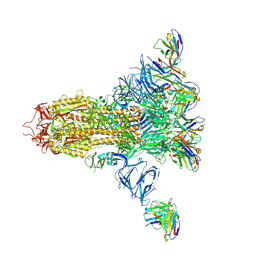 | |
7LY0
 
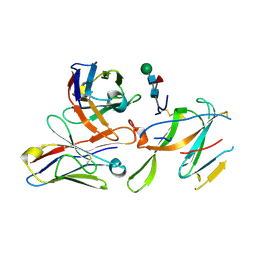 | |
7LY2
 
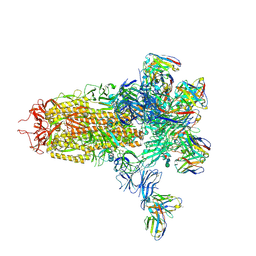 | |
7LY3
 
 | |
7ME0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 | | 分子名称: | Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | 登録日 | 2021-04-06 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.48 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | 分子名称: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | 著者 | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2020-10-04 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | 分子名称: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | 著者 | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2020-10-04 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7DX4
 
 | | The structure of FC08 Fab-hA.CE2-RBD complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Heavy chain of FC08 Fab, ... | | 著者 | Cao, L, Wang, X. | | 登録日 | 2021-01-18 | | 公開日 | 2021-04-21 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
7KRN
 
 | | Structure of SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | 著者 | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | 登録日 | 2020-11-20 | | 公開日 | 2021-04-21 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|








