9UHT
 
 | | SARS-CoV-2 E-RTC in complex with RNA-nsp9 and GMPPNP | | Descriptor: | Helicase nsp13, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Huang, Y.C, Liu, Y.X, Lou, Z.Y, Rao, Z.H, Yan, L.M. | | Deposit date: | 2025-04-14 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Response to Matter Arising: Substrate selection and transition state support the mechanism for GTP-mediate RNA capping catalyzed by SARS-CoV-2 NiRAN
To Be Published
|
|
9C7X
 
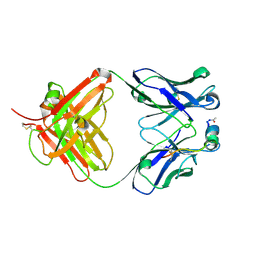 | |
9EEI
 
 | |
9EET
 
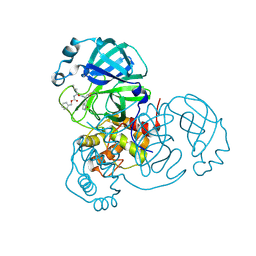 | |
9EEV
 
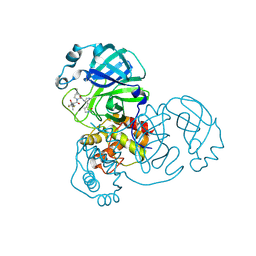 | | Crystal structure of the SARS-CoV-2 Omicron nsp5 main protease (Mpro) E166V mutant in complex with inhibitor Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Neilsen, G, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2024-11-19 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Strategy to overcome a nirmatrelvir resistance mechanism in the SARS-CoV-2 nsp5 protease.
Sci Adv, 11, 2025
|
|
9FMW
 
 | | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6 | | Descriptor: | Fab of neutralizing mAb K501SP6 heavy chain, Fab of neutralizing mAb K501SP6 light chain, Spike glycoprotein,Fibritin | | Authors: | Bjoernsson, K.H, Walker, M.R, Raghavan, S.S.R, Ward, A.B, Barfod, L.K. | | Deposit date: | 2024-06-07 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Omicron BA.1 Spike protein with neutralizing NTD specific mAb K501SP6
To Be Published
|
|
9IKZ
 
 | | SARS-CoV-2 E-RTC bound to pRNA-nsp9 and GDP-BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Helicase nsp13, ... | | Authors: | Yan, L.M, Huang, Y.C, Liu, Y.X, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2024-06-29 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Substrate selection and transition reveal the mechanism for RNA capping catalyzed by SARS-CoV-2 NiRAN
To Be Published
|
|
9VAO
 
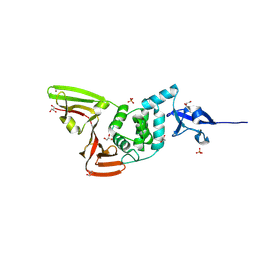 | | Crystal structure of Papain-like protease (PLpro) from SARS-CoV-2 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Papain-like protease nsp3, ... | | Authors: | Arya, R, Ganesh, J, Prashar, V, Kumar, M. | | Deposit date: | 2025-06-03 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of Papain-like protease (PLpro) from SARS-CoV-2
To Be Published
|
|
9CPP
 
 | |
9CPQ
 
 | |
9CPR
 
 | |
9CPS
 
 | |
9CPT
 
 | |
9CPU
 
 | |
9CPV
 
 | |
9CPW
 
 | |
9CPX
 
 | |
9CPY
 
 | |
9P0F
 
 | |
9CFE
 
 | |
9CFF
 
 | |
9CFG
 
 | |
9CFH
 
 | |
9G0H
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the noncovalently bound inhibitor C5N17A | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, C.-C, Strater, N, Sylvester, K, Muller, C.E, Yang, M, Lee, M.K, Gao, S, Song, L, Liu, X, Kim, M, Zhan, P. | | Deposit date: | 2024-07-08 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Miniaturized Modular Click Chemistry-enabled Rapid Discovery of Unique SARS-CoV-2 M pro Inhibitors With Robust Potency and Drug-like Profile.
Adv Sci, 11, 2024
|
|
9G0I
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the noncovalently bound inhibitor C5N17B | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, IMIDAZOLE, ... | | Authors: | Yang, C.-C, Strater, N, Sylvester, K, Muller, C.E, Yang, M, Lee, M.K, Gao, S, Song, L, Liu, X, Kim, M, Zhan, P. | | Deposit date: | 2024-07-08 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Miniaturized Modular Click Chemistry-enabled Rapid Discovery of Unique SARS-CoV-2 M pro Inhibitors With Robust Potency and Drug-like Profile.
Adv Sci, 11, 2024
|
|








