8Z9L
 
 | |
8ZBF
 
 | |
9DTZ
 
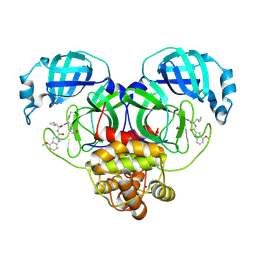 | | SARS-CoV-2 Mpro in complex with compound 5 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclopropyl-1-({(2R)-1-imino-3-[(3R)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]-4-methoxy-1H-indole-2-carboxamide, SODIUM ION | | Authors: | Bigelow, L, Tang, H, Boguslaw, N, Duda, D.M. | | Deposit date: | 2024-10-02 | | Release date: | 2025-04-30 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of dimeric SARS-CoV-2 Mpro displays positive cooperativity and a mixture of covalent and non-covalent binding.
Iscience, 28, 2025
|
|
9DU2
 
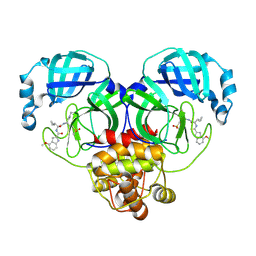 | | SARS-CoV-2 Mpro in complex with compound 7 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclopropyl-1-{[(1Z,2S)-1-imino-4-(methanesulfonyl)butan-2-yl]amino}-1-oxopropan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Bigelow, L, Tang, H, Boguslaw, N, Duda, D.M. | | Deposit date: | 2024-10-02 | | Release date: | 2025-04-30 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inhibition of dimeric SARS-CoV-2 Mpro displays positive cooperativity and a mixture of covalent and non-covalent binding.
Iscience, 28, 2025
|
|
9DU3
 
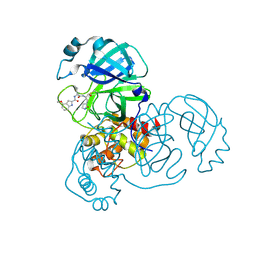 | | SARS-CoV-2 Mpro in complex with compound 1 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclopropyl-1-({(2R)-1-hydroxy-3-[(3R)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Bigelow, L, Tang, H, Boguslaw, N, Duda, D.M. | | Deposit date: | 2024-10-02 | | Release date: | 2025-04-30 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Inhibition of dimeric SARS-CoV-2 Mpro displays positive cooperativity and a mixture of covalent and non-covalent binding.
Iscience, 28, 2025
|
|
9DU4
 
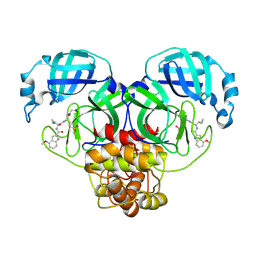 | | SARS-CoV-2 Mpro in complex with compound 3 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclopropyl-1-{[(2R)-1-hydroxy-4-(methanesulfonyl)butan-2-yl]amino}-1-oxopropan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Bigelow, L, Tang, H, Boguslaw, N, Duda, D.M. | | Deposit date: | 2024-10-02 | | Release date: | 2025-04-30 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Inhibition of dimeric SARS-CoV-2 Mpro displays positive cooperativity and a mixture of covalent and non-covalent binding.
Iscience, 28, 2025
|
|
8WZ0
 
 | | SARS-CoV-2 3CLpro bound to covalent inhibitor | | Descriptor: | (2~{R})-~{N}-[4,4-bis(fluoranyl)cyclohexyl]-2-[(2-chloranyl-2-fluoranyl-ethanoyl)-[4-(trifluoromethyloxy)phenyl]amino]-2-pyrimidin-5-yl-propanamide, 3C-like proteinase nsp5 | | Authors: | Peng, J, Sun, D, Ding, X. | | Deposit date: | 2023-11-01 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | SARS-CoV-2 3CLpro bound to covalent inhibitor
To Be Published
|
|
9BF8
 
 | | SARS-CoV-2 Papain-like Protease (PLpro) Untagged Crystal Structure | | Descriptor: | 1,2-ETHANEDIOL, ORF1a polyprotein, PHOSPHATE ION, ... | | Authors: | Al-Homoudi, A.I, Engel, J, Brunzelle, J.S, Gavande, N, Kovari, L.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human Structural Homologues of SARS-CoV-2 PL pro as Anti-Targets: A Strategic Panel Analysis.
MicroPubl Biol, 2025, 2025
|
|
9FM4
 
 | | Dynamic structure of the apical stem loop of the stem loop 2 motif (s2m) from SCoV-2 Delta variant | | Descriptor: | RNA (25-MER) | | Authors: | Wirtz Martin, M.A, Matzel, T, Makowski, J, Kensinger, A, Herr, A, Wacker, A, Richter, C, Jonker, H.R.A, Evanseck, J, Schwalbe, H. | | Deposit date: | 2024-06-05 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-16 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structural heterogeneity and dynamics in the apical stem loop of s2m from SARS-CoV-2 Delta by an integrative NMR spectroscopy and MD simulation approach.
Nucleic Acids Res., 53, 2025
|
|
8X1X
 
 | | SARS-CoV-2 Papain like protease (PLpro) in complex with inhibitor Lithocholic acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, GLYCEROL, Papain-like protease nsp3, ... | | Authors: | Choudhary, S, Verma, S, Kumar, P, Tomar, S. | | Deposit date: | 2023-11-09 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SARS-CoV-2 Papain like protease (PLpro) in complex with inhibitor Lithocholic acid
To Be Published
|
|
9BLF
 
 | | SARS-CoV-2 core polymerase complex inhibited by araCTP | | Descriptor: | 4-amino-1-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}pyrimidin-2(1H)-one, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Anderson, T.K, Kirchdeorfer, R.N. | | Deposit date: | 2024-04-30 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure of the SARS-CoV-2 core-RTC bound to RNA, araCMP, and araCTP
To Be Published
|
|
9BQF
 
 | |
9BQG
 
 | |
9BQL
 
 | |
9BQM
 
 | |
9BQN
 
 | |
9BQO
 
 | |
9BQP
 
 | |
9BQQ
 
 | |
9BQT
 
 | |
9BQY
 
 | |
9BQZ
 
 | |
9BR0
 
 | |
9BR1
 
 | |
9BS7
 
 | |








