[English] 日本語
 Yorodumi
Yorodumi- PDB-7lhz: K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA an... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7lhz | ||||||
|---|---|---|---|---|---|---|---|
| Title | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid (compound 25) | ||||||
 Components Components |
| ||||||
 Keywords Keywords | ISOMERASE/DNA/INHIBITOR / Gyrase / DNA topoisomerase / ParC / ParE / ISOMERASE-DNA-INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationDNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity / DNA topoisomerase (ATP-hydrolysing) / DNA topological change / chromosome segregation / chromosome / DNA binding / ATP binding Similarity search - Function | ||||||
| Biological species |  Klebsiella pneumoniae 342 (bacteria) Klebsiella pneumoniae 342 (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 3.3 Å molecular replacement / Resolution: 3.3 Å | ||||||
 Authors Authors | Noeske, J. / Shu, W. / Bellamacina, C. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2021 Journal: J.Med.Chem. / Year: 2021Title: Discovery and Optimization of DNA Gyrase and Topoisomerase IV Inhibitors with Potent Activity against Fluoroquinolone-Resistant Gram-Positive Bacteria. Authors: Lapointe, G. / Skepper, C.K. / Holder, L.M. / Armstrong, D. / Bellamacina, C. / Blais, J. / Bussiere, D. / Bian, J. / Cepura, C. / Chan, H. / Dean, C.R. / De Pascale, G. / Dhumale, B. / ...Authors: Lapointe, G. / Skepper, C.K. / Holder, L.M. / Armstrong, D. / Bellamacina, C. / Blais, J. / Bussiere, D. / Bian, J. / Cepura, C. / Chan, H. / Dean, C.R. / De Pascale, G. / Dhumale, B. / Fisher, L.M. / Fulsunder, M. / Kantariya, B. / Kim, J. / King, S. / Kossy, L. / Kulkarni, U. / Lakshman, J. / Leeds, J.A. / Ling, X. / Lvov, A. / Ma, S. / Malekar, S. / McKenney, D. / Mergo, W. / Metzger, L. / Mhaske, K. / Moser, H.E. / Mostafavi, M. / Namballa, S. / Noeske, J. / Osborne, C. / Patel, A. / Patel, D. / Patel, T. / Piechon, P. / Polyakov, V. / Prajapati, K. / Prosen, K.R. / Reck, F. / Richie, D.L. / Sanderson, M.R. / Satasia, S. / Savani, B. / Selvarajah, J. / Sethuraman, V. / Shu, W. / Tashiro, K. / Thompson, K.V. / Vaarla, K. / Vala, L. / Veselkov, D.A. / Vo, J. / Vora, B. / Wagner, T. / Wedel, L. / Williams, S.L. / Yendluri, S. / Yue, Q. / Yifru, A. / Zhang, Y. / Rivkin, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7lhz.cif.gz 7lhz.cif.gz | 1.4 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7lhz.ent.gz pdb7lhz.ent.gz | 1.2 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7lhz.json.gz 7lhz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lh/7lhz https://data.pdbj.org/pub/pdb/validation_reports/lh/7lhz ftp://data.pdbj.org/pub/pdb/validation_reports/lh/7lhz ftp://data.pdbj.org/pub/pdb/validation_reports/lh/7lhz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5eixS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 4 molecules CDAB
| #1: Protein | Mass: 84199.500 Da / Num. of mol.: 4 Fragment: UNP residues 390-631 (parE) + UNP residues 1-490 (parC) Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Klebsiella pneumoniae 342 (bacteria) / Strain: 342 / Gene: parE, NCTC5052_03997, parC, SAMEA4873570_03328 / Production host: Klebsiella pneumoniae 342 (bacteria) / Strain: 342 / Gene: parE, NCTC5052_03997, parC, SAMEA4873570_03328 / Production host:  References: UniProt: A0A377Y395, UniProt: A0A486EJ79, DNA topoisomerase (ATP-hydrolysing) |
|---|
-DNA chain , 2 types, 8 molecules PJHEKIGF
| #2: DNA chain | Mass: 3354.206 Da / Num. of mol.: 4 / Source method: obtained synthetically / Source: (synth.)  Klebsiella pneumoniae 342 (bacteria) Klebsiella pneumoniae 342 (bacteria)#3: DNA chain | Mass: 4586.025 Da / Num. of mol.: 4 / Source method: obtained synthetically / Source: (synth.)  Klebsiella pneumoniae 342 (bacteria) Klebsiella pneumoniae 342 (bacteria) |
|---|
-Non-polymers , 4 types, 14 molecules 

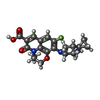




| #4: Chemical | | #5: Chemical | ChemComp-MG / #6: Chemical | ChemComp-Y21 / ( #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.96 Å3/Da / Density % sol: 58.45 % |
|---|---|
| Crystal grow | Temperature: 290 K / Method: vapor diffusion, sitting drop / pH: 6.6 Details: 20% MPD, 10 mM magnesium chloride, 0.5 mM spermine, 100 mM sodium cacodylate, pH 6.6 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 17-ID / Wavelength: 1 Å / Beamline: 17-ID / Wavelength: 1 Å | |||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Mar 9, 2017 | |||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 | |||||||||||||||||||||
| Reflection | Resolution: 3.3→54.91 Å / Num. obs: 63836 / % possible obs: 99.6 % / Redundancy: 3.3 % / CC1/2: 0.979 / Rmerge(I) obs: 0.247 / Net I/σ(I): 3.3 | |||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 5EIX Resolution: 3.3→46.92 Å / SU ML: 0.6 / Cross valid method: THROUGHOUT / σ(F): 1.33 / Phase error: 33.9 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 222.3 Å2 / Biso mean: 83.2138 Å2 / Biso min: 18.78 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3.3→46.92 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 23
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj








































