+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5lty | ||||||
|---|---|---|---|---|---|---|---|
| Title | Homeobox transcription factor CDX2 bound to methylated DNA | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSCRIPTION / transcription factor / methylated DNA | ||||||
| Function / homology |  Function and homology information Function and homology informationintestinal epithelial cell differentiation / regulation of somitogenesis / trophectodermal cell differentiation / establishment or maintenance of epithelial cell apical/basal polarity / embryonic pattern specification / labyrinthine layer development / methyl-CpG binding / digestive tract development / anterior/posterior axis specification / endosome to lysosome transport ...intestinal epithelial cell differentiation / regulation of somitogenesis / trophectodermal cell differentiation / establishment or maintenance of epithelial cell apical/basal polarity / embryonic pattern specification / labyrinthine layer development / methyl-CpG binding / digestive tract development / anterior/posterior axis specification / endosome to lysosome transport / blood vessel development / somatic stem cell population maintenance / transcription repressor complex / animal organ morphogenesis / stem cell differentiation / condensed nuclear chromosome / RNA polymerase II transcription regulatory region sequence-specific DNA binding / positive regulation of cell differentiation / DNA-binding transcription repressor activity, RNA polymerase II-specific / sequence-specific double-stranded DNA binding / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / DNA-binding transcription factor activity, RNA polymerase II-specific / cell differentiation / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / positive regulation of cell population proliferation / regulation of transcription by RNA polymerase II / chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / nucleoplasm / nucleus Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.66 Å MOLECULAR REPLACEMENT / Resolution: 2.66 Å | ||||||
 Authors Authors | Morgunova, E. / Popov, A. / Taipale, J. | ||||||
 Citation Citation |  Journal: Science / Year: 2017 Journal: Science / Year: 2017Title: Impact of cytosine methylation on DNA binding specificities of human transcription factors. Authors: Yin, Y. / Morgunova, E. / Jolma, A. / Kaasinen, E. / Sahu, B. / Khund-Sayeed, S. / Das, P.K. / Kivioja, T. / Dave, K. / Zhong, F. / Nitta, K.R. / Taipale, M. / Popov, A. / Ginno, P.A. / ...Authors: Yin, Y. / Morgunova, E. / Jolma, A. / Kaasinen, E. / Sahu, B. / Khund-Sayeed, S. / Das, P.K. / Kivioja, T. / Dave, K. / Zhong, F. / Nitta, K.R. / Taipale, M. / Popov, A. / Ginno, P.A. / Domcke, S. / Yan, J. / Schubeler, D. / Vinson, C. / Taipale, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5lty.cif.gz 5lty.cif.gz | 154.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5lty.ent.gz pdb5lty.ent.gz | 120.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5lty.json.gz 5lty.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lt/5lty https://data.pdbj.org/pub/pdb/validation_reports/lt/5lty ftp://data.pdbj.org/pub/pdb/validation_reports/lt/5lty ftp://data.pdbj.org/pub/pdb/validation_reports/lt/5lty | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5ef6C  5egoC  5hodC  5luxC  5eeaS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 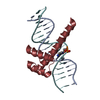
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 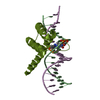
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: _ / Refine code: _
NCS ensembles :
|
- Components
Components
| #1: DNA chain | Mass: 5462.551 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) Homo sapiens (human)#2: Protein | Mass: 8869.227 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CDX2, CDX3 / Plasmid: pETG20A / Production host: Homo sapiens (human) / Gene: CDX2, CDX3 / Plasmid: pETG20A / Production host:  #3: DNA chain | Mass: 5596.684 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)  Homo sapiens (human) Homo sapiens (human)#4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.4 Å3/Da / Density % sol: 48.73 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 8 Details: PEG 5000, potassium chloride, magnesium chloride, PEG 400,Tris buffer |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-1 / Wavelength: 0.9724 Å / Beamline: ID23-1 / Wavelength: 0.9724 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Jul 2, 2016 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9724 Å / Relative weight: 1 |
| Reflection | Resolution: 2.66→63.29 Å / Num. obs: 10979 / % possible obs: 98.5 % / Redundancy: 3.3 % / Biso Wilson estimate: 73.25 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.043 / Net I/σ(I): 15 |
| Reflection shell | Resolution: 2.66→2.79 Å / Redundancy: 3.1 % / Rmerge(I) obs: 0.637 / Mean I/σ(I) obs: 1.7 / CC1/2: 0.647 / % possible all: 96.7 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5EEA Resolution: 2.66→63.29 Å / Cor.coef. Fo:Fc: 0.961 / Cor.coef. Fo:Fc free: 0.932 / SU B: 31.14 / SU ML: 0.293 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R Free: 0.337 Details: U VALUES : WITH TLS ADDED HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 201.37 Å2 / Biso mean: 77.843 Å2 / Biso min: 36.77 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.66→63.29 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Refine-ID: X-RAY DIFFRACTION / Type: interatomic distance / Weight position: 0.05
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.656→2.725 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller









 PDBj
PDBj










































