[English] 日本語
 Yorodumi
Yorodumi- PDB-4v5q: The crystal structure of EF-Tu and G24A-tRNA-Trp bound to a near-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4v5q | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The crystal structure of EF-Tu and G24A-tRNA-Trp bound to a near- cognate codon on the 70S ribosome | ||||||||||||
 Components Components |
| ||||||||||||
 Keywords Keywords | RIBOSOME / HIRSH TRNA | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationtranslation elongation factor activity / large ribosomal subunit / regulation of translation / ribosomal small subunit assembly / small ribosomal subunit / 5S rRNA binding / large ribosomal subunit rRNA binding / transferase activity / cytosolic small ribosomal subunit / ribosomal large subunit assembly ...translation elongation factor activity / large ribosomal subunit / regulation of translation / ribosomal small subunit assembly / small ribosomal subunit / 5S rRNA binding / large ribosomal subunit rRNA binding / transferase activity / cytosolic small ribosomal subunit / ribosomal large subunit assembly / cytosolic large ribosomal subunit / tRNA binding / negative regulation of translation / rRNA binding / ribosome / structural constituent of ribosome / ribonucleoprotein complex / translation / GTPase activity / mRNA binding / GTP binding / RNA binding / zinc ion binding / metal ion binding / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |   THERMUS THERMOPHILUS (bacteria) THERMUS THERMOPHILUS (bacteria) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.1 Å MOLECULAR REPLACEMENT / Resolution: 3.1 Å | ||||||||||||
 Authors Authors | Schmeing, T.M. / Voorhees, R.M. / Ramakrishnan, V. | ||||||||||||
 Citation Citation |  Journal: Nat.Struct.Mol.Biol. / Year: 2011 Journal: Nat.Struct.Mol.Biol. / Year: 2011Title: How Mutations in tRNA Distant from the Anticodon Affect the Fidelity of Decoding. Authors: Schmeing, T.M. / Voorhees, R.M. / Kelley, A.C. / Ramakrishnan, V. | ||||||||||||
| History |
| ||||||||||||
| Remark 700 | SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "QA" IN EACH CHAIN ON SHEET RECORDS BELOW ... SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "QA" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 5-STRANDED BARREL THIS IS REPRESENTED BY A 6-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4v5q.cif.gz 4v5q.cif.gz | 7.5 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4v5q.ent.gz pdb4v5q.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  4v5q.json.gz 4v5q.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4v5q_validation.pdf.gz 4v5q_validation.pdf.gz | 3.3 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4v5q_full_validation.pdf.gz 4v5q_full_validation.pdf.gz | 5.3 MB | Display | |
| Data in XML |  4v5q_validation.xml.gz 4v5q_validation.xml.gz | 912.7 KB | Display | |
| Data in CIF |  4v5q_validation.cif.gz 4v5q_validation.cif.gz | 1.2 MB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v5/4v5q https://data.pdbj.org/pub/pdb/validation_reports/v5/4v5q ftp://data.pdbj.org/pub/pdb/validation_reports/v5/4v5q ftp://data.pdbj.org/pub/pdb/validation_reports/v5/4v5q | HTTPS FTP |
-Related structure data
| Related structure data |  4v5pC  4v5rC  4v5sC  2wrn  2jl5  2jl7 C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-RNA chain , 6 types, 14 molecules AACAAVAWCVCWAXCXAYCYBADABBDB
| #1: RNA chain | Mass: 493958.281 Da / Num. of mol.: 2 / Source method: isolated from a natural source Details: CHAIN A (16S RNA) HAS E.COLI NUMBERING, BASED ON A STRUCTURAL ALIGNMENT WITH THE CORRESPONDING E. COLI STRUCTURE IN 2AVY. Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: GenBank: 55979969 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: GenBank: 55979969#22: RNA chain | Mass: 24485.539 Da / Num. of mol.: 4 / Source method: isolated from a natural source / Source: (natural)  #23: RNA chain | Mass: 8808.352 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.)  #24: RNA chain | Mass: 24813.127 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: MUTATION AT G24A / Source: (natural)  #36: RNA chain | Mass: 947975.500 Da / Num. of mol.: 2 / Source method: isolated from a natural source Details: CHAIN A (23S RNA) HAS E.COLI RESIDUE NUMBERING, BASED ON A STRUCTURAL ALIGNMENT WITH THE CORRESPONDING E. COLI STRUCTURE IN 2AW4 Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: GenBank: 55979969 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: GenBank: 55979969#37: RNA chain | Mass: 39540.617 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: GenBank: 55979969 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: GenBank: 55979969 |
|---|
-30S RIBOSOMAL PROTEIN ... , 20 types, 40 molecules ABCBACCCADCDAECEAFCFAGCGAHCHAICIAJCJAKCKALCLAMCMANCNAOCOAPCP...
| #2: Protein | Mass: 29317.703 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80371 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80371#3: Protein | Mass: 26751.076 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80372 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80372#4: Protein | Mass: 24373.447 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80373 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80373#5: Protein | Mass: 17583.416 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHQ5 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHQ5#6: Protein | Mass: 11988.753 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SLP8 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SLP8#7: Protein | Mass: 18050.973 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P17291 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P17291#8: Protein | Mass: 15868.570 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHQ2, UniProt: A0A0M9AFS9*PLUS THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHQ2, UniProt: A0A0M9AFS9*PLUS#9: Protein | Mass: 14410.614 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80374 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80374#10: Protein | Mass: 11954.968 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHN7 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHN7#11: Protein | Mass: 13737.868 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80376 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80376#12: Protein | Mass: 14637.384 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHN3 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHN3#13: Protein | Mass: 14338.861 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80377 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80377#14: Protein | Mass: 7158.725 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHQ1, UniProt: A0A0N0BLP2*PLUS THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHQ1, UniProt: A0A0N0BLP2*PLUS#15: Protein | Mass: 10578.407 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SJ76 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SJ76#16: Protein | Mass: 10409.983 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SJH3 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SJH3#17: Protein | Mass: 12325.655 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHP7, UniProt: A0A0N0BLS5*PLUS THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHP7, UniProt: A0A0N0BLS5*PLUS#18: Protein | Mass: 10258.299 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SLQ0 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SLQ0#19: Protein | Mass: 10605.464 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHP2 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SHP2#20: Protein | Mass: 11736.143 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80380 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: P80380#21: Protein/peptide | Mass: 3350.030 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SIH3 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 - MRC - MSAW1 / References: UniProt: Q5SIH3 |
|---|
-Protein , 1 types, 2 molecules AZCZ
| #25: Protein | Mass: 44709.852 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   THERMUS THERMOPHILUS (bacteria) / Strain: HB8 / References: UniProt: Q5SHN6 THERMUS THERMOPHILUS (bacteria) / Strain: HB8 / References: UniProt: Q5SHN6 |
|---|
+50S RIBOSOMAL PROTEIN ... , 31 types, 62 molecules B0D0B1D1B2D2B3D3B4D4B5D5B6D6B7D7B8D8B9D9BCDCBDDDBEDEBFDFBGDG...
-Non-polymers , 3 types, 12 molecules 

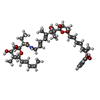


| #59: Chemical | ChemComp-ZN / #60: Chemical | #61: Chemical | |
|---|
-Details
| Has protein modification | Y |
|---|---|
| Sequence details | G24A MUTATION |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.39 Å3/Da / Density % sol: 63.67 % / Description: NONE |
|---|---|
| Crystal grow | pH: 6.3 Details: 100 MM MES PH 6.3, 60-100 MM KCL, 50 MM SUCROSE, 1% GLYCEROL, 5.2% (W/V) PEG20K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 0.9535 / Beamline: ID14-4 / Wavelength: 0.9535 |
| Detector | Type: ADSC / Detector: CCD / Date: Jan 28, 2010 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9535 Å / Relative weight: 1 |
| Reflection | Resolution: 3.1→50 Å / Num. obs: 1105502 / % possible obs: 98.7 % / Observed criterion σ(I): 1.27 / Redundancy: 6.4 % / Biso Wilson estimate: 61.1 Å2 / Rmerge(I) obs: 0.02 / Net I/σ(I): 6.19 |
| Reflection shell | Resolution: 3.1→3.2 Å / Redundancy: 6.5 % / Rmerge(I) obs: 0.09 / Mean I/σ(I) obs: 1.23 / % possible all: 98.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2WRN  2wrn Resolution: 3.1→50 Å / Rfactor Rfree error: 0.001 / Data cutoff high absF: 13186381 / Data cutoff low absF: 0 / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 45.2195 Å2 / ksol: 0.3 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 90.7 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.1→50 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | NCS model details: RESTRAINTS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.1→3.29 Å / Rfactor Rfree error: 0.004 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller

























































































 PDBj
PDBj
































