[English] 日本語
 Yorodumi
Yorodumi- PDB-1fka: STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1fka | ||||||
|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RIBOSOME / 30S RIBOSOMAL SUBUNIT / PROTEIN-RNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / tRNA binding / rRNA binding / structural constituent of ribosome / ribosome / translation ...ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / tRNA binding / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / metal ion binding / cytoplasm Similarity search - Function | ||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 3.3 Å SYNCHROTRON / Resolution: 3.3 Å | ||||||
 Authors Authors | Schluenzen, F. / Tocilj, A. / Zarivach, R. / Harms, J. / Gluehmann, M. / Janell, D. / Bashan, A. / Bartels, H. / Agmon, I. / Franceschi, F. / Yonath, A. | ||||||
 Citation Citation |  Journal: Cell(Cambridge,Mass.) / Year: 2000 Journal: Cell(Cambridge,Mass.) / Year: 2000Title: Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Authors: Schluenzen, F. / Tocilj, A. / Zarivach, R. / Harms, J. / Gluehmann, M. / Janell, D. / Bashan, A. / Bartels, H. / Agmon, I. / Franceschi, F. / Yonath, A. #1:  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: Refining the overall structure and subdomain orientation of ribosomal protein S4 delta 41 with dipolar couplings measured by nmr in uniaxial liquid crystalline phases Authors: MARKUS, M.A. / GERSTNER, R.B. / DRAPER, D.E. / Torchia, D.A. #2:  Journal: Nature / Year: 1992 Journal: Nature / Year: 1992Title: The structure of ribosomal protein s5 reveals sites of interaction with 16S rRNA Authors: RAMAKRISHNAN, V. / WHITE, S.W. #3:  Journal: Embo J. / Year: 1994 Journal: Embo J. / Year: 1994Title: Crystal structure of the ribosomal protein S6 from Thermus thermophilus Authors: LINDAHL, M. / SVENSSON, L.A. / LILJAS, A. / SEDELNIKOVA, S.E. / ELISEIKINA, I.A. / FOMENKOVA, N.P. / NEVSKAYA, N. / NIKONOV, S.V. / GARBER, M.B. / MURANOVA, T.A. / RYKONOVA, A.I. #4:  Journal: Structure / Year: 1997 Journal: Structure / Year: 1997Title: The structure of ribosomal protein S7 at 1.9 A resolution reveals a beta-hairpin motif that binds double-stranded nucleic acids Authors: WIMBERLY, B.T. / WHITE, S.W. / RAMAKRISHNAN, V. #5:  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: Crystal structure of ribosomal protein S8 from Thermus thermophilus reveals a high degree of structural conservation of a specific RNA binding site Authors: NEVSKAYA, N. / TISHCHENKO, S. / NIKULIN, A. / AL-KARADAGHI, S. / LILJAS, A. / EHRESMANN, B. / EHRESMANN, C. / GARBER, M.B. / NIKONOV, S. #6:  Journal: Structure / Year: 1998 Journal: Structure / Year: 1998Title: Conformational variability of the N-terminal helix in the structure of ribosomal protein S15 Authors: CLEMONS, W.M. / DAVIES, C. / WHITE, S.W. / RAMAKRISHNAN, V. #7:  Journal: Science / Year: 2000 Journal: Science / Year: 2000Title: Structure of the S15, S6, S18-rRNA complex: assembly of the 30S ribosome central domain Authors: AGALAROV, S.C. / PRASAD, G.S. / FUNKE, P.M. / STOUT, C.D. / WILLIAMSON, J.R. #8:  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: Solution structure of the ribosomal protein S19 from Thermus thermophilus Authors: HELGSTRAND, M. / RAK, A.V. / ALLARD, P. / DAVYDOVA, N. / GARBER, M.B. / HARD, T. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1fka.cif.gz 1fka.cif.gz | 819.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1fka.ent.gz pdb1fka.ent.gz | 582.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1fka.json.gz 1fka.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fk/1fka https://data.pdbj.org/pub/pdb/validation_reports/fk/1fka ftp://data.pdbj.org/pub/pdb/validation_reports/fk/1fka ftp://data.pdbj.org/pub/pdb/validation_reports/fk/1fka | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
-30S RIBOSOMAL PROTEIN ... , 19 types, 19 molecules BCDEFGHIJKLMNOPQRST
| #2: Protein | Mass: 9464.658 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
|---|---|
| #3: Protein | Mass: 14996.461 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #4: Protein | Mass: 24373.447 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P80373 Thermus thermophilus (bacteria) / References: UniProt: P80373 |
| #5: Protein | Mass: 17583.416 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P27152, UniProt: Q5SHQ5*PLUS Thermus thermophilus (bacteria) / References: UniProt: P27152, UniProt: Q5SHQ5*PLUS |
| #6: Protein | Mass: 11988.753 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P23370, UniProt: Q5SLP8*PLUS Thermus thermophilus (bacteria) / References: UniProt: P23370, UniProt: Q5SLP8*PLUS |
| #7: Protein | Mass: 17335.170 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P17291 Thermus thermophilus (bacteria) / References: UniProt: P17291 |
| #8: Protein | Mass: 15868.570 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P24319, UniProt: P0DOY9*PLUS Thermus thermophilus (bacteria) / References: UniProt: P24319, UniProt: P0DOY9*PLUS |
| #9: Protein | Mass: 7592.350 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #10: Protein | Mass: 6060.462 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #11: Protein | Mass: 5975.357 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #12: Protein | Mass: 8783.818 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #13: Protein | Mass: 6571.091 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #14: Protein/peptide | Mass: 2230.741 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #15: Protein | Mass: 10622.417 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P80378, UniProt: Q5SJ76*PLUS Thermus thermophilus (bacteria) / References: UniProt: P80378, UniProt: Q5SJ76*PLUS |
| #16: Protein | Mass: 6230.672 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #17: Protein | Mass: 7166.825 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #18: Protein | Mass: 10244.272 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: GenBank: 6739549, UniProt: Q5SLQ0*PLUS Thermus thermophilus (bacteria) / References: GenBank: 6739549, UniProt: Q5SLQ0*PLUS |
| #19: Protein | Mass: 10605.464 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P80381, UniProt: Q5SHP2*PLUS Thermus thermophilus (bacteria) / References: UniProt: P80381, UniProt: Q5SHP2*PLUS |
| #20: Protein | Mass: 8102.979 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
-RNA chain / Non-polymers , 2 types, 8 molecules A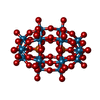

| #1: RNA chain | Mass: 492673.531 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: GenBank: 155076 Thermus thermophilus (bacteria) / References: GenBank: 155076 |
|---|---|
| #21: Chemical | ChemComp-WO2 / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 5.14 Å3/Da / Density % sol: 76.09 % | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 7.7 Details: MPD, Spermidine, MgCl2, NH4Cl, pH 7.7, VAPOR DIFFUSION, HANGING DROP, temperature 291.0K | ||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||
| Crystal grow | *PLUS Details: Tsiboli, P., (1994) Eur. J. Biochem., 226, 169. | ||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 95 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-2 / Wavelength: 0.933 / Beamline: ID14-2 / Wavelength: 0.933 |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Jun 15, 2000 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.933 Å / Relative weight: 1 |
| Reflection | Resolution: 3.3→35 Å / Num. all: 206724 / Num. obs: 206724 / % possible obs: 94.7 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 9.8 % / Rmerge(I) obs: 0.136 / Net I/σ(I): 21.9 |
| Reflection shell | Resolution: 3.3→3.52 Å / Redundancy: 2.5 % / Rmerge(I) obs: 0.441 / % possible all: 77.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 3.3→35 Å / Cross valid method: THROUGHOUT / σ(F): 3 / σ(I): 0 / Stereochemistry target values: none
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.3→35 Å
| |||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Version: 0.9 / Classification: refinement | |||||||||||||||||||||||||
| Refinement | *PLUS σ(F): 3 / % reflection Rfree: 10 % | |||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller




















 PDBj
PDBj






























