+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1qd7 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
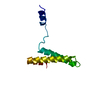
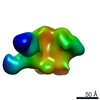
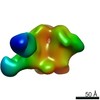
| Title | PARTIAL MODEL FOR 30S RIBOSOMAL SUBUNIT | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | RIBOSOME / 30S RIBOSOMAL SUBUNIT / LOW RESOLUTION MODEL | |||||||||
| Function / homology |  Function and homology information Function and homology informationribosomal small subunit biogenesis / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / tRNA binding / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex ...ribosomal small subunit biogenesis / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / tRNA binding / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / response to antibiotic / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 5.5 Å MAD / Resolution: 5.5 Å | |||||||||
 Authors Authors | Clemons Jr., W.M. / May, J.L.C. / Wimberly, B.T. / McCutcheon, J.P. / Capel, M.S. / Ramakrishnan, V. | |||||||||
 Citation Citation |  Journal: Nature / Year: 1999 Journal: Nature / Year: 1999Title: Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution. Authors: Clemons Jr., W.M. / May, J.L. / Wimberly, B.T. / McCutcheon, J.P. / Capel, M.S. / Ramakrishnan, V. #1:  Journal: Embo J. / Year: 1998 Journal: Embo J. / Year: 1998Title: The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: one domain shows structural homology to the ETS DNA-binding motif Authors: Davies, C. / Gerstner, R.B. / Draper, D.E. / Ramakrishnan, V. / White, S.W. #2:  Journal: Nature / Year: 1992 Journal: Nature / Year: 1992Title: The structure of ribosomal protein S5 reveals sites of interaction with 16S rRNA Authors: Ramakrishnan, V. / White, S.W. #3:  Journal: Embo J. / Year: 1994 Journal: Embo J. / Year: 1994Title: Crystal structure of the ribosomal protein S6 from Thermus thermophilus Authors: Lindahl, M. / Svensson, L.A. / Liljas, A. / Sedelnikova, I.A. / Eliseikina, I.A. / Fomenkova, N.P. / Nevskaya, N. / Nikonov, S.V. / Garber, M.B. / Muranova, T.A. / Rykonova, A.I. / Amons, R. #4:  Journal: Structure / Year: 1997 Journal: Structure / Year: 1997Title: The structure of ribosomal protein S7 at 1.9 A resolution reveals a beta- hairpin motif that binds double-stranded nucleic acids Authors: Wimberly, B.T. / White, S.W. / Ramakrishnan, V. #5:  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: Crystal structure of ribosomal protein S8 from Thermus thermophilus reveals a high degree of structural conservation of a specific RNA binding site Authors: Nevskaya, N. / Tischenko, S. / Nikulin, A. / Al-Karadaghi, S. / Liljas, A. / Ehresmann, B. / Ehresmann, C. / Garber, M. / Nikonov, S. #6:  Journal: Structure / Year: 1998 Journal: Structure / Year: 1998Title: Conformational variability of the N-terminal helix in the structure of ribosomal protein S15 Authors: Clemons Jr., W.M. / Davies, C.R. / White, S.W. / Ramakrishnan, V. #7:  Journal: Biochemistry / Year: 1996 Journal: Biochemistry / Year: 1996Title: Solution structure of prokaryotic ribosomal protein S17 by high-resolution NMR spectroscopy Authors: Jaishree, T.N. / Ramakrishnan, V. / White, S.W. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1qd7.cif.gz 1qd7.cif.gz | 60.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1qd7.ent.gz pdb1qd7.ent.gz | 33.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1qd7.json.gz 1qd7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qd/1qd7 https://data.pdbj.org/pub/pdb/validation_reports/qd/1qd7 ftp://data.pdbj.org/pub/pdb/validation_reports/qd/1qd7 ftp://data.pdbj.org/pub/pdb/validation_reports/qd/1qd7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1a23S  1an7S  1pkpS  1risS  1rssS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
-RNA chain , 2 types, 2 molecules AB
| #1: RNA chain | Mass: 53096.625 Da / Num. of mol.: 1 / Fragment: RESIDUES 563-912 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
|---|---|
| #2: RNA chain | Mass: 17211.391 Da / Num. of mol.: 1 / Fragment: RESIDUES 1400-1500 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
-Protein , 8 types, 8 molecules CDEFGHIJ
| #3: Protein | Mass: 18624.359 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P81288 Thermus thermophilus (bacteria) / References: UniProt: P81288 |
|---|---|
| #4: Protein | Mass: 15240.724 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P02357 Thermus thermophilus (bacteria) / References: UniProt: P02357 |
| #5: Protein | Mass: 11619.337 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P23370, UniProt: Q5SLP8*PLUS Thermus thermophilus (bacteria) / References: UniProt: P23370, UniProt: Q5SLP8*PLUS |
| #6: Protein | Mass: 15342.846 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P17291 Thermus thermophilus (bacteria) / References: UniProt: P17291 |
| #7: Protein | Mass: 15624.214 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P24319, UniProt: P0DOY9*PLUS Thermus thermophilus (bacteria) / References: UniProt: P24319, UniProt: P0DOY9*PLUS |
| #8: Protein | Mass: 10200.844 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P05766 Thermus thermophilus (bacteria) / References: UniProt: P05766 |
| #9: Protein | Mass: 10081.853 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P23828 Thermus thermophilus (bacteria) / References: UniProt: P23828 |
| #10: Protein | Mass: 8528.504 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 6 X-RAY DIFFRACTION / Number of used crystals: 6 |
|---|
- Sample preparation
Sample preparation
| Crystal grow | Temperature: 277 K / Method: vapor diffusion / Details: VAPOUR DIFFUSION AT 277 K, MPD, VAPOR DIFFUSION | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Components of the solutions |
| ||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop / Details: Trakhanov, S.D., (1987) FEBS Lett., 220, 319. | ||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X25 / Wavelength: 1.000, 1.700 / Beamline: X25 / Wavelength: 1.000, 1.700 | |||||||||
| Detector | Type: BRANDEIS - B4 / Detector: CCD / Date: Mar 4, 1999 | |||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||
| Radiation wavelength |
| |||||||||
| Reflection | Highest resolution: 5.5 Å / Num. all: 42000 / Num. obs: 42000 / % possible obs: 95 % / Observed criterion σ(I): 0 / Redundancy: 5 % | |||||||||
| Reflection shell | Highest resolution: 5.5 Å / Mean I/σ(I) obs: 5.1 / % possible all: 83 | |||||||||
| Reflection | *PLUS |
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD MADStarting model: PROTEINS WERE FIT INTO MAP AS RIGID BODIES FROM KNOWN CRYSTAL OR NMR STRUCTURES OF ISOLATED PROTEINS EXCEPT FOR S20, FOR WHICH NO HIGH RESOLUTION STRUCTURE EXISTS. S20 WAS FIT AS ...Starting model: PROTEINS WERE FIT INTO MAP AS RIGID BODIES FROM KNOWN CRYSTAL OR NMR STRUCTURES OF ISOLATED PROTEINS EXCEPT FOR S20, FOR WHICH NO HIGH RESOLUTION STRUCTURE EXISTS. S20 WAS FIT AS THREE-HELIX BUNDLE.S4 SEE REFERENCE (1); S5 MODELED ACCORDING TO PDB CODE 1PKP;S6 MODELED ACCORDING TO PDB CODE 1RIS;S7 MODELED ACCORDING TO PDB CODE 1RSS;S8 MODELED ACCORDING TO PDB CODE 1AN7;S15 MODELED ACCORDING TO PDB CODE 1A23;S17 SEE REFERENCE (7); Highest resolution: 5.5 Å / Num. reflection all: 42000 / Num. reflection obs: 42000 Details: NO REFINEMENT WAS DONE EXCEPT FOR VISUAL MAP FITTING. THIS MODEL WAS MADE FROM A 5.5 ANGSTROM X-RAY MAP BY FITTING A-FORM RNA HELICES INTO REGIONS OF DOUBLE- HELICAL DENSITY. NO ATTEMPT HAS ...Details: NO REFINEMENT WAS DONE EXCEPT FOR VISUAL MAP FITTING. THIS MODEL WAS MADE FROM A 5.5 ANGSTROM X-RAY MAP BY FITTING A-FORM RNA HELICES INTO REGIONS OF DOUBLE- HELICAL DENSITY. NO ATTEMPT HAS BEEN MADE TO MAINTAIN PROPER STEREOCHEMISTRY OR EVEN PHOSPHATE-PHOSPHATE DISTANCES. IN PARTICULAR AT JUNCTIONS OF THE SHORT HELICES, THE CHAIN MAY BE UNREALISTIC | ||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 5.5 Å
| ||||||||||||
| Refinement | *PLUS Highest resolution: 5.5 Å | ||||||||||||
| Solvent computation | *PLUS | ||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller









 PDBj
PDBj





























