+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3g2d | ||||||
|---|---|---|---|---|---|---|---|
| Title | Complex of Mth0212 and a 4 bp dsDNA with 3'-overhang | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE/DNA / protein-DNA complex / dsDNA with 3'-overhang / HYDROLASE-DNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationexodeoxyribonuclease III / double-stranded DNA 3'-5' DNA exonuclease activity / phosphoric diester hydrolase activity / class I DNA-(apurinic or apyrimidinic site) endonuclease activity / DNA-(apurinic or apyrimidinic site) lyase / base-excision repair / endonuclease activity / DNA binding / metal ion binding Similarity search - Function | ||||||
| Biological species |   Methanothermobacter thermautotrophicus (archaea) Methanothermobacter thermautotrophicus (archaea) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.3 Å molecular replacement / Resolution: 2.3 Å | ||||||
 Authors Authors | Lakomek, K. / Dickmanns, A. / Ficner, R. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2010 Journal: J.Mol.Biol. / Year: 2010Title: Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA Authors: Lakomek, K. / Dickmanns, A. / Ciirdaeva, E. / Schomacher, L. / Ficner, R. #1: Journal: Nucleic Acids Res. / Year: 2006 Title: The Methanothermobacter thermautotrophicus ExoIII homologue Mth212 is a DNA uridine endonuclease Authors: Georg, J. / Schomacher, L. / Chong, J.P.J. / Majernik, A.I. / Raabe, M. / Urlaub, H. / Muller, S. / Ciirdaeva, E. / Kramer, W. / Fritz, H.-J. | ||||||
| History |
| ||||||
| Remark 285 | CRYST1 THE DEPOSITOR CONFIRM THE REPORTED SPACE GROUP P 1 21 1 DESPITE OF THE BETA ANGLE OF 90.0 ...CRYST1 THE DEPOSITOR CONFIRM THE REPORTED SPACE GROUP P 1 21 1 DESPITE OF THE BETA ANGLE OF 90.0 DEGREE. THE STRUCTURE COULD NOT BE REFINED IN A PRIMITIVE ORTHORHOMBIC SPACE GROUP - MOST LIKELY DUE TO SOME DISORDER IN THE CRYSTAL OR TWINNING. HOWEVER, THERE WAS NO INDICATION OF TWINNING. THE PRESENCE OF PEG MOLECULES IN ONLY ONE OF THE TWO PROTEIN MOLECULES IN THE ASYMMETRIC UNIT (ASU) AT EQUIVALENT POSITIONS (IN SPACE GROUP P21) AND DIFFERENCES IN THE DOUBLE-STRANDED DNA HELICES (DIFFERENT LENGTH OF ONE STRAND) BOUND BY THE TWO MOLECULES IN THE ASU E.G. ARGUED AGAINST MODEL BUILDING IN AN ORTHORHOMBIC SPACE GROUP. HOWEVER, THE USE OF A TWIN LAW MIGHT HAVE BEEN AN ADVANTAGE, BUT WAS NOT TAKEN INTO ACCOUNT DUE TO THE RESOLUTION OF 2.3A PREVENTING REFINEMENT WITH SHELXL AND DNA RESTRAINTS IN CNS LESS SUITABLE THAN THE RESTRAINTS USED BY REFMAC5 FOR EXAMPLE. THE STRUCTURE COULD BE REFINED TO REASONABLE R VALUES EVEN WITHOUT THE APPLICATION OF A TWIN LAW. THUS, THE REFINEMENT WAS CONTINUED IN THE SPACE GROUP P21. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3g2d.cif.gz 3g2d.cif.gz | 145.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3g2d.ent.gz pdb3g2d.ent.gz | 110 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3g2d.json.gz 3g2d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g2/3g2d https://data.pdbj.org/pub/pdb/validation_reports/g2/3g2d ftp://data.pdbj.org/pub/pdb/validation_reports/g2/3g2d ftp://data.pdbj.org/pub/pdb/validation_reports/g2/3g2d | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3fziSC  3g00C  3g0aC  3g0rC  3g1kC  3g2cC  3g38C  3g3cC  3g3yC  3g4tC  3g8vC  3g91C  3ga6C S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 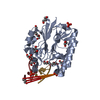
| ||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: 1 / Beg auth comp-ID: ALA / Beg label comp-ID: ALA / Refine code: 1
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 31429.666 Da / Num. of mol.: 2 / Mutation: T2A, D151N Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Methanothermobacter thermautotrophicus (archaea) Methanothermobacter thermautotrophicus (archaea)Strain: Delta H (DSM 1053) / Gene: mth0212, MTH212, MTH_212 / Plasmid: pET_B_001-mth212 (D151N) / Production host:  |
|---|
-DNA chain , 2 types, 4 molecules GHKI
| #2: DNA chain | Mass: 2741.800 Da / Num. of mol.: 2 / Source method: obtained synthetically #3: DNA chain | Mass: 3021.964 Da / Num. of mol.: 2 / Source method: obtained synthetically |
|---|
-Non-polymers , 4 types, 327 molecules 






| #4: Chemical | | #5: Chemical | ChemComp-PG4 / #6: Chemical | ChemComp-GOL / #7: Water | ChemComp-HOH / | |
|---|
-Details
| Compound details | THE 3'-OVERHANG OF THE DNA COMPRISES 4 AND 5 NUCLEOTIDES, RESPECTIVELY, ADJACENT TO THE TWO DOUBLE- ...THE 3'-OVERHANG OF THE DNA COMPRISES 4 AND 5 NUCLEOTIDE |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.37 Å3/Da / Density % sol: 48.02 % | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 5.8 Details: reservoir: 5% (w/v) PEG 4000, 50mM KCl, 50mM MES pH 5.8, 10mM MgCl2 ; complex solution: 240mM NaCl, 8mM HEPES-KOH pH 7.6, 4mM DTT, 2mM MgCl2, 1mM KH2PO4/K2HPO4 pH 7.0, VAPOR DIFFUSION, ...Details: reservoir: 5% (w/v) PEG 4000, 50mM KCl, 50mM MES pH 5.8, 10mM MgCl2 ; complex solution: 240mM NaCl, 8mM HEPES-KOH pH 7.6, 4mM DTT, 2mM MgCl2, 1mM KH2PO4/K2HPO4 pH 7.0, VAPOR DIFFUSION, SITTING DROP, temperature 293K | ||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X13 / Wavelength: 0.815 Å / Beamline: X13 / Wavelength: 0.815 Å |
| Detector | Type: MAR CCD 165 mm / Detector: CCD / Date: Mar 27, 2007 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.815 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→50 Å / Num. obs: 29120 / % possible obs: 93.8 % / Redundancy: 2.9 % / Rsym value: 0.055 / Net I/σ(I): 18 |
| Reflection shell | Resolution: 2.3→2.38 Å / Redundancy: 2.6 % / Mean I/σ(I) obs: 4.2 / Rsym value: 0.191 / % possible all: 76.3 |
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 3FZI Resolution: 2.3→48.55 Å / Cor.coef. Fo:Fc: 0.921 / Cor.coef. Fo:Fc free: 0.862 / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.787 / SU B: 7.971 / SU ML: 0.198 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.445 / ESU R Free: 0.289 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 107.75 Å2 / Biso mean: 28.354 Å2 / Biso min: 9.4 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→48.55 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Dom-ID: 1 / Auth asym-ID: A / Ens-ID: 1 / Number: 2122 / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.296→2.356 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller













 PDBj
PDBj





