+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31116 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
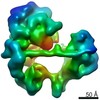
| Title | TFIID lobe B subcomplex | |||||||||
 Map data Map data | TFIID lobe B subcomplex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TFIID / preinitiation complex / core promoter / transcription initiation / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationSAGA complex assembly / lateral mesodermal cell differentiation / DNA-templated transcription open complex formation / allantois development / pre-snoRNP complex / transcription factor TFTC complex / SLIK (SAGA-like) complex / hepatocyte differentiation / positive regulation of response to cytokine stimulus / maintenance of protein location in nucleus ...SAGA complex assembly / lateral mesodermal cell differentiation / DNA-templated transcription open complex formation / allantois development / pre-snoRNP complex / transcription factor TFTC complex / SLIK (SAGA-like) complex / hepatocyte differentiation / positive regulation of response to cytokine stimulus / maintenance of protein location in nucleus / C2H2 zinc finger domain binding / RNA polymerase binding / regulation of fat cell differentiation / limb development / box C/D snoRNP assembly / SAGA complex / transcription preinitiation complex / inner cell mass cell proliferation / RNA polymerase II general transcription initiation factor activity / negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / transcription factor TFIID complex / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / regulation of RNA splicing / negative regulation of cell cycle / aryl hydrocarbon receptor binding / MLL1 complex / RNA polymerase II transcribes snRNA genes / positive regulation of transcription initiation by RNA polymerase II / embryonic placenta development / somitogenesis / regulation of DNA repair / RNA polymerase II preinitiation complex assembly / ovarian follicle development / positive regulation of intrinsic apoptotic signaling pathway / RNA Polymerase II Pre-transcription Events / negative regulation of proteasomal ubiquitin-dependent protein catabolic process / response to interleukin-1 / TBP-class protein binding / nuclear estrogen receptor binding / male germ cell nucleus / transcription initiation at RNA polymerase II promoter / promoter-specific chromatin binding / DNA-templated transcription initiation / mRNA transcription by RNA polymerase II / G1/S transition of mitotic cell cycle / multicellular organism growth / p53 binding / actin cytoskeleton / HATs acetylate histones / ATPase binding / Regulation of TP53 Activity through Phosphorylation / DNA-binding transcription factor binding / transcription by RNA polymerase II / transcription coactivator activity / cell differentiation / transcription cis-regulatory region binding / Ub-specific processing proteases / protein stabilization / positive regulation of apoptotic process / chromatin remodeling / protein heterodimerization activity / negative regulation of cell population proliferation / apoptotic process / DNA damage response / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process / chromatin / positive regulation of DNA-templated transcription / perinuclear region of cytoplasm / enzyme binding / positive regulation of transcription by RNA polymerase II / protein-containing complex / DNA binding / nucleoplasm / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.77 Å | |||||||||
 Authors Authors | Chen X / Wu Z | |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Structural insights into preinitiation complex assembly on core promoters. Authors: Xizi Chen / Yilun Qi / Zihan Wu / Xinxin Wang / Jiabei Li / Dan Zhao / Haifeng Hou / Yan Li / Zishuo Yu / Weida Liu / Mo Wang / Yulei Ren / Ze Li / Huirong Yang / Yanhui Xu /  Abstract: Transcription factor IID (TFIID) recognizes core promoters and supports preinitiation complex (PIC) assembly for RNA polymerase II (Pol II)-mediated eukaryotic transcription. We determined the ...Transcription factor IID (TFIID) recognizes core promoters and supports preinitiation complex (PIC) assembly for RNA polymerase II (Pol II)-mediated eukaryotic transcription. We determined the structures of human TFIID-based PIC in three stepwise assembly states and revealed two-track PIC assembly: stepwise promoter deposition to Pol II and extensive modular reorganization on track I (on TATA-TFIID-binding element promoters) versus direct promoter deposition on track II (on TATA-only and TATA-less promoters). The two tracks converge at an ~50-subunit holo PIC in identical conformation, whereby TFIID stabilizes PIC organization and supports loading of cyclin-dependent kinase (CDK)-activating kinase (CAK) onto Pol II and CAK-mediated phosphorylation of the Pol II carboxyl-terminal domain. Unexpectedly, TBP of TFIID similarly bends TATA box and TATA-less promoters in PIC. Our study provides structural visualization of stepwise PIC assembly on highly diversified promoters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31116.map.gz emd_31116.map.gz | 64.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31116-v30.xml emd-31116-v30.xml emd-31116.xml emd-31116.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31116.png emd_31116.png | 140.3 KB | ||
| Filedesc metadata |  emd-31116.cif.gz emd-31116.cif.gz | 7.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31116 http://ftp.pdbj.org/pub/emdb/structures/EMD-31116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31116 | HTTPS FTP |
-Related structure data
| Related structure data |  7eggMC  7edxC  7eg7C  7eg8C  7eg9C  7egaC  7egbC  7egcC  7egdC  7egeC  7egfC  7eghC  7egiC  7egjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31116.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31116.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TFIID lobe B subcomplex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TFIID lobe B subcomplex
| Entire | Name: TFIID lobe B subcomplex |
|---|---|
| Components |
|
-Supramolecule #1: TFIID lobe B subcomplex
| Supramolecule | Name: TFIID lobe B subcomplex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transcription initiation factor TFIID subunit 4
| Macromolecule | Name: Transcription initiation factor TFIID subunit 4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 110.221883 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAGSDLLDE VFFNSEVDEK VVSDLVGSLE SQLAASAAHH HHLAPRTPEV RAAAAGALGN HVVSGSPAGA AGAGPAAPAE GAPGAAPEP PPAGRARPGG GGPQRPGPPS PRRPLVPAGP APPAAKLRPP PEGSAGSCAP VPAAAAVAAG PEPAPAGPAK P AGPAALAA ...String: MAAGSDLLDE VFFNSEVDEK VVSDLVGSLE SQLAASAAHH HHLAPRTPEV RAAAAGALGN HVVSGSPAGA AGAGPAAPAE GAPGAAPEP PPAGRARPGG GGPQRPGPPS PRRPLVPAGP APPAAKLRPP PEGSAGSCAP VPAAAAVAAG PEPAPAGPAK P AGPAALAA RAGPGPGPGP GPGPGPGPGK PAGPGAAQTL NGSAALLNSH HAAAPAVSLV NNGPAALLPL PKPAAPGTVI QT PPFVGAA APPAPAAPSP PAAPAPAAPA AAPPPPPPAP ATLARPPGHP AGPPTAAPAV PPPAAAQNGG SAGAAPAPAP AAG GPAGVS GQPGPGAAAA APAPGVKAES PKRVVQAAPP AAQTLAASGP ASTAASMVIG PTMQGALPSP AAVPPPAPGT PTGL PKGAA GAVTQSLSRT PTATTSGIRA TLTPTVLAPR LPQPPQNPTN IQNFQLPPGM VLVRSENGQL LMIPQQALAQ MQAQA HAQP QTTMAPRPAT PTSAPPVQIS TVQAPGTPII ARQVTPTTII KQVSQAQTTV QPSATLQRSP GVQPQLVLGG AAQTAS LGT ATAVQTGTPQ RTVPGATTTS SAATETMENV KKCKNFLSTL IKLASSGKQS TETAANVKEL VQNLLDGKIE AEDFTSR LY RELNSSPQPY LVPFLKRSLP ALRQLTPDSA AFIQQSQQQP PPPTSQATTA LTAVVLSSSV QRTAGKTAAT VTSALQPP V LSLTQPTQVG VGKQGQPTPL VIQQPPKPGA LIRPPQVTLT QTPMVALRQP HNRIMLTTPQ QIQLNPLQPV PVVKPAVLP GTKALSAVSA QAAAAQKNKL KEPGGGSFRD DDDINDVASM AGVNLSEESA RILATNSELV GTLTRSCKDE TFLLQAPLQR RILEIGKKH GITELHPDVV SYVSHATQQR LQNLVEKISE TAQQKNFSYK DDDRYEQASD VRAQLKFFEQ LDQIEKQRKD E QEREILMR AAKSRSRQED PEQLRLKQKA KEMQQQELAQ MRQRDANLTA LAAIGPRKKR KVDCPGPGSG AEGSGPGSVV PG SSGVGTP RQFTRQRITR VNLRDLIFCL ENERETSHSL LLYKAFLK UniProtKB: Transcription initiation factor TFIID subunit 4 |
-Macromolecule #2: Transcription initiation factor TFIID subunit 5
| Macromolecule | Name: Transcription initiation factor TFIID subunit 5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 86.932109 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAALAEEQTE VAVKLEPEGP PTLLPPQAGD GAGEGSGGTT NNGPNGGGGN VAASSSTGGD GGTPKPTVAV SAAAPAGAAP VPAAAPDAG APHDRQTLLA VLQFLRQSKL REAEEALRRE AGLLEEAVAG SGAPGEVDSA GAEVTSALLS RVTASAPGPA A PDPPGTGA ...String: MAALAEEQTE VAVKLEPEGP PTLLPPQAGD GAGEGSGGTT NNGPNGGGGN VAASSSTGGD GGTPKPTVAV SAAAPAGAAP VPAAAPDAG APHDRQTLLA VLQFLRQSKL REAEEALRRE AGLLEEAVAG SGAPGEVDSA GAEVTSALLS RVTASAPGPA A PDPPGTGA SGATVVSGSA SGPAAPGKVG SVAVEDQPDV SAVLSAYNQQ GDPTMYEEYY SGLKHFIECS LDCHRAELSQ LF YPLFVHM YLELVYNQHE NEAKSFFEKF HGDQECYYQD DLRVLSSLTK KEHMKGNETM LDFRTSKFVL RISRDSYQLL KRH LQEKQN NQIWNIVQEH LYIDIFDGMP RSKQQIDAMV GSLAGEAKRE ANKSKVFFGL LKEPEIEVPL DDEDEEGENE EGKP KKKKP KKDSIGSKSK KQDPNAPPQN RIPLPELKDS DKLDKIMNMK ETTKRVRLGP DCLPSICFYT FLNAYQGLTA VDVTD DSSL IAGGFADSTV RVWSVTPKKL RSVKQASDLS LIDKESDDVL ERIMDEKTAS ELKILYGHSG PVYGASFSPD RNYLLS SSE DGTVRLWSLQ TFTCLVGYKG HNYPVWDTQF SPYGYYFVSG GHDRVARLWA TDHYQPLRIF AGHLADVNCT RFHPNSN YV ATGSADRTVR LWDVLNGNCV RIFTGHKGPI HSLTFSPNGR FLATGATDGR VLLWDIGHGL MVGELKGHTD TVCSLRFS R DGEILASGSM DNTVRLWDAI KAFEDLETDD FTTATGHINL PENSQELLLG TYMTKSTPVV HLHFTRRNLV LAAGAYSPQ UniProtKB: Transcription initiation factor TFIID subunit 5 |
-Macromolecule #3: Transcription initiation factor TFIID subunit 6
| Macromolecule | Name: Transcription initiation factor TFIID subunit 6 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.749297 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAEEKKLKLS NTVLPSESMK VVAESMGIAQ IQEETCQLLT DEVSYRIKEI AQDALKFMHM GKRQKLTTSD IDYALKLKNV EPLYGFHAQ EFIPFRFASG GGRELYFYEE KEVDLSDIIN TPLPRVPLDV CLKAHWLSIE GCQPAIPENP PPAPKEQQKA E ATEPLKSA ...String: MAEEKKLKLS NTVLPSESMK VVAESMGIAQ IQEETCQLLT DEVSYRIKEI AQDALKFMHM GKRQKLTTSD IDYALKLKNV EPLYGFHAQ EFIPFRFASG GGRELYFYEE KEVDLSDIIN TPLPRVPLDV CLKAHWLSIE GCQPAIPENP PPAPKEQQKA E ATEPLKSA KPGQEEDGPL KGKGQGATTA DGKGKEKKAP PLLEGAPLRL KPRSIHELSV EQQLYYKEIT EACVGSCEAK RA EALQSIA TDPGLYQMLP RFSTFISEGV RVNVVQNNLA LLIYLMRMVK ALMDNPTLYL EKYVHELIPA VMTCIVSRQL CLR PDVDNH WALRDFAARL VAQICKHFST TTNNIQSRIT KTFTKSWVDE KTPWTTRYGS IAGLAELGHD VIKTLILPRL QQEG ERIRS VLDGPVLSNI DRIGADHVQS LLLKHCAPVL AKLRPPPDNQ DAYRAEFGSL GPLLCSQVVK ARAQAALQAQ QVNRT TLTI TQPRPTLTLS QAPQPGPRTP GLLKVPGSIA LPVQTLVSAR AAAPPQPSPP PTKFIVMSSS SSAPSTQQVL SLSTSA PGS GSTTTSPVTT TVPSVQPIVK LVSTATTAPP STAPSGPGSV QKYIVVSLPP TGEGKGGPTS HPSPVPPPAS SPSPLSG SA LCGGKQEAGD SPPPAPGTPK ANGSQPNSGS PQPAP UniProtKB: Transcription initiation factor TFIID subunit 6 |
-Macromolecule #4: Transcription initiation factor TFIID subunit 8
| Macromolecule | Name: Transcription initiation factor TFIID subunit 8 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.304359 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADAAATAGA GGSGTRSGSK QSTNPADNYH LARRRTLQVV VSSLLTEAGF ESAEKASVET LTEMLQSYIS EIGRSAKSYC EHTARTQPT LSDIVVTLVE MGFNVDTLPA YAKRSQRMVI TAPPVTNQPV TPKALTAGQN RPHPPHIPSH FPEFPDPHTY I KTPTYREP ...String: MADAAATAGA GGSGTRSGSK QSTNPADNYH LARRRTLQVV VSSLLTEAGF ESAEKASVET LTEMLQSYIS EIGRSAKSYC EHTARTQPT LSDIVVTLVE MGFNVDTLPA YAKRSQRMVI TAPPVTNQPV TPKALTAGQN RPHPPHIPSH FPEFPDPHTY I KTPTYREP VSDYQVLREK AASQRRDVER ALTRFMAKTG ETQSLFKDDV STFPLIAARP FTIPYLTALL PSELEMQQME ET DSSEQDE QTDTENLALH ISMEDSGAEK ENTSVLQQNP SLSGSRNGEE NIIDNPYLRP VKKPKIRRKK SLS UniProtKB: Transcription initiation factor TFIID subunit 8 |
-Macromolecule #5: Transcription initiation factor TFIID subunit 9
| Macromolecule | Name: Transcription initiation factor TFIID subunit 9 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.006838 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MESGKTASPK SMPKDAQMMA QILKDMGITE YEPRVINQML EFAFRYVTTI LDDAKIYSSH AKKATVDADD VRLAIQCRAD QSFTSPPPR DFLLDIARQR NQTPLPLIKP YSGPRLPPDR YCLTAPNYRL KSLQKKASTS AGRITVPRLS VGSVTSRPST P TLGTPTPQ ...String: MESGKTASPK SMPKDAQMMA QILKDMGITE YEPRVINQML EFAFRYVTTI LDDAKIYSSH AKKATVDADD VRLAIQCRAD QSFTSPPPR DFLLDIARQR NQTPLPLIKP YSGPRLPPDR YCLTAPNYRL KSLQKKASTS AGRITVPRLS VGSVTSRPST P TLGTPTPQ TMSVSTKVGT PMSLTGQRFT VQMPTSQSPA VKASIPATSA VQNVLINPSL IGSKNILITT NMMSSQNTAN ES SNALKRK REDDDDDDDD DDDYDNL UniProtKB: Transcription initiation factor TFIID subunit 9 |
-Macromolecule #6: Transcription initiation factor TFIID subunit 10
| Macromolecule | Name: Transcription initiation factor TFIID subunit 10 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 21.731248 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSCSGSGADP EAAPASAASA PGPAPPVSAP AALPSSTAAE NKASPAGTAG GPGAGAAAGG TGPLAARAGE PAERRGAAPV SAGGAAPPE GAISNGVYVL PSAANGDVKP VVSSTPLVDF LMQLEDYTPT IPDAVTGYYL NRAGFEASDP RIIRLISLAA Q KFISDIAN ...String: MSCSGSGADP EAAPASAASA PGPAPPVSAP AALPSSTAAE NKASPAGTAG GPGAGAAAGG TGPLAARAGE PAERRGAAPV SAGGAAPPE GAISNGVYVL PSAANGDVKP VVSSTPLVDF LMQLEDYTPT IPDAVTGYYL NRAGFEASDP RIIRLISLAA Q KFISDIAN DALQHCKMKG TASGSSRSKS KDRKYTLTME DLTPALSEYG INVKKPHYFT UniProtKB: Transcription initiation factor TFIID subunit 10 |
-Macromolecule #7: Transcription initiation factor TFIID subunit 12
| Macromolecule | Name: Transcription initiation factor TFIID subunit 12 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.948467 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNQFGPSALI NLSNFSSIKP EPASTPPQGS MANSTAVVKI PGTPGAGGRL SPENNQVLTK KKLQDLVREV DPNEQLDEDV EEMLLQIAD DFIESVVTAA CQLARHRKSS TLEVKDVQLH LERQWNMWIP GFGSEEIRPY KKACTTEAHK QRMALIRKTT K K UniProtKB: Transcription initiation factor TFIID subunit 12 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Grid | Material: GOLD / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 63.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.77 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 84427 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7egg: |
 Movie
Movie Controller
Controller














































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















