+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31117 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
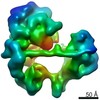
| Title | TFIID lobe C subcomplex | |||||||||
 Map data Map data | TFIID lobe C subcomplex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TFIID / preinitiation complex / core promoter / transcription initiation / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of MHC class I biosynthetic process / response to paraquat / spermine transport / DNA-templated transcription open complex formation / positive regulation of androgen receptor signaling pathway / TFIIH-class transcription factor complex binding / negative regulation of protein autoubiquitination / negative regulation of MHC class II biosynthetic process / transcription factor TFTC complex / regulation of cell cycle G1/S phase transition ...negative regulation of MHC class I biosynthetic process / response to paraquat / spermine transport / DNA-templated transcription open complex formation / positive regulation of androgen receptor signaling pathway / TFIIH-class transcription factor complex binding / negative regulation of protein autoubiquitination / negative regulation of MHC class II biosynthetic process / transcription factor TFTC complex / regulation of cell cycle G1/S phase transition / RNA polymerase I general transcription initiation factor activity / SLIK (SAGA-like) complex / maintenance of protein location in nucleus / RNA polymerase II general transcription initiation factor binding / nuclear vitamin D receptor binding / regulation of fat cell differentiation / nuclear thyroid hormone receptor binding / SAGA complex / inner cell mass cell proliferation / RNA polymerase II general transcription initiation factor activity / transcription factor TFIID complex / histone acetyltransferase binding / midbrain development / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / cellular response to ATP / negative regulation of signal transduction by p53 class mediator / aryl hydrocarbon receptor binding / negative regulation of cell cycle / transcription initiation at RNA polymerase I promoter / histone acetyltransferase activity / ubiquitin conjugating enzyme activity / P-TEFb complex binding / MLL1 complex / RNA polymerase II transcribes snRNA genes / histone H4K16ac reader activity / negative regulation of ubiquitin-dependent protein catabolic process / positive regulation of transcription initiation by RNA polymerase II / RNA polymerase II core promoter sequence-specific DNA binding / regulation of DNA repair / negative regulation of protein kinase activity / RNA polymerase II preinitiation complex assembly / transcription regulator inhibitor activity / histone acetyltransferase / estrogen receptor signaling pathway / positive regulation of intrinsic apoptotic signaling pathway / RNA Polymerase II Pre-transcription Events / TBP-class protein binding / regulation of signal transduction by p53 class mediator / nuclear receptor binding / male germ cell nucleus / transcription initiation at RNA polymerase II promoter / DNA-templated transcription initiation / mRNA transcription by RNA polymerase II / G2/M transition of mitotic cell cycle / protein polyubiquitination / p53 binding / kinase activity / cellular response to UV / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / protein autophosphorylation / chromatin organization / transcription regulator complex / sequence-specific DNA binding / Regulation of TP53 Activity through Phosphorylation / DNA-binding transcription factor binding / ubiquitin-dependent protein catabolic process / transcription by RNA polymerase II / RNA polymerase II-specific DNA-binding transcription factor binding / transcription coactivator activity / cell differentiation / protein kinase activity / non-specific serine/threonine protein kinase / transcription cis-regulatory region binding / protein stabilization / positive regulation of apoptotic process / protein heterodimerization activity / negative regulation of cell population proliferation / negative regulation of gene expression / protein serine kinase activity / negative regulation of DNA-templated transcription / protein serine/threonine kinase activity / apoptotic process / DNA damage response / chromatin binding / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / chromatin / perinuclear region of cytoplasm / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / protein-containing complex / DNA binding / nucleoplasm / ATP binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Chen X / Wu Z | |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Structural insights into preinitiation complex assembly on core promoters. Authors: Xizi Chen / Yilun Qi / Zihan Wu / Xinxin Wang / Jiabei Li / Dan Zhao / Haifeng Hou / Yan Li / Zishuo Yu / Weida Liu / Mo Wang / Yulei Ren / Ze Li / Huirong Yang / Yanhui Xu /  Abstract: Transcription factor IID (TFIID) recognizes core promoters and supports preinitiation complex (PIC) assembly for RNA polymerase II (Pol II)-mediated eukaryotic transcription. We determined the ...Transcription factor IID (TFIID) recognizes core promoters and supports preinitiation complex (PIC) assembly for RNA polymerase II (Pol II)-mediated eukaryotic transcription. We determined the structures of human TFIID-based PIC in three stepwise assembly states and revealed two-track PIC assembly: stepwise promoter deposition to Pol II and extensive modular reorganization on track I (on TATA-TFIID-binding element promoters) versus direct promoter deposition on track II (on TATA-only and TATA-less promoters). The two tracks converge at an ~50-subunit holo PIC in identical conformation, whereby TFIID stabilizes PIC organization and supports loading of cyclin-dependent kinase (CDK)-activating kinase (CAK) onto Pol II and CAK-mediated phosphorylation of the Pol II carboxyl-terminal domain. Unexpectedly, TBP of TFIID similarly bends TATA box and TATA-less promoters in PIC. Our study provides structural visualization of stepwise PIC assembly on highly diversified promoters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31117.map.gz emd_31117.map.gz | 3.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31117-v30.xml emd-31117-v30.xml emd-31117.xml emd-31117.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31117.png emd_31117.png | 107 KB | ||
| Filedesc metadata |  emd-31117.cif.gz emd-31117.cif.gz | 8.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31117 http://ftp.pdbj.org/pub/emdb/structures/EMD-31117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31117 | HTTPS FTP |
-Related structure data
| Related structure data |  7eghMC  7edxC  7eg7C  7eg8C  7eg9C  7egaC  7egbC  7egcC  7egdC  7egeC  7egfC  7eggC  7egiC  7egjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31117.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31117.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TFIID lobe C subcomplex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.055 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TFIID lobe C subcomplex
| Entire | Name: TFIID lobe C subcomplex |
|---|---|
| Components |
|
-Supramolecule #1: TFIID lobe C subcomplex
| Supramolecule | Name: TFIID lobe C subcomplex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transcription initiation factor TFIID subunit 1
| Macromolecule | Name: Transcription initiation factor TFIID subunit 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: histone acetyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 212.956172 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGPGCDLLLR TAATITAAAI MSDTDSDEDS AGGGPFSLAG FLFGNINGAG QLEGESVLDD ECKKHLAGLG ALGLGSLITE LTANEELTG TDGALVNDEG WVRSTEDAVD YSDINEVAED ESRRYQQTMG SLQPLCHSDY DEDDYDADCE DIDCKLMPPP P PPPGPMKK ...String: MGPGCDLLLR TAATITAAAI MSDTDSDEDS AGGGPFSLAG FLFGNINGAG QLEGESVLDD ECKKHLAGLG ALGLGSLITE LTANEELTG TDGALVNDEG WVRSTEDAVD YSDINEVAED ESRRYQQTMG SLQPLCHSDY DEDDYDADCE DIDCKLMPPP P PPPGPMKK DKDQDSITGE KVDFSSSSDS ESEMGPQEAT QAESEDGKLT LPLAGIMQHD ATKLLPSVTE LFPEFRPGKV LR FLRLFGP GKNVPSVWRS ARRKRKKKHR ELIQEEQIQE VECSVESEVS QKSLWNYDYA PPPPPEQCLS DDEITMMAPV ESK FSQSTG DIDKVTDTKP RVAEWRYGPA RLWYDMLGVP EDGSGFDYGF KLRKTEHEPV IKSRMIEEFR KLEENNGTDL LADE NFLMV TQLHWEDDII WDGEDVKHKG TKPQRASLAG WLPSSMTRNA MAYNVQQGFA ATLDDDKPWY SIFPIDNEDL VYGRW EDNI IWDAQAMPRL LEPPVLTLDP NDENLILEIP DEKEEATSNS PSKESKKESS LKKSRILLGK TGVIKEEPQQ NMSQPE VKD PWNLSNDEYY YPKQQGLRGT FGGNIIQHSI PAVELRQPFF PTHMGPIKLR QFHRPPLKKY SFGALSQPGP HSVQPLL KH IKKKAKMREQ ERQASGGGEM FFMRTPQDLT GKDGDLILAE YSEENGPLMM QVGMATKIKN YYKRKPGKDP GAPDCKYG E TVYCHTSPFL GSLHPGQLLQ AFENNLFRAP IYLHKMPETD FLIIRTRQGY YIRELVDIFV VGQQCPLFEV PGPNSKRAN THIRDFLQVF IYRLFWKSKD RPRRIRMEDI KKAFPSHSES SIRKRLKLCA DFKRTGMDSN WWVLKSDFRL PTEEEIRAMV SPEQCCAYY SMIAAEQRLK DAGYGEKSFF APEEENEEDF QMKIDDEVRT APWNTTRAFI AAMKGKCLLE VTGVADPTGC G EGFSYVKI PNKPTQQKDD KEPQPVKKTV TGTDADLRRL SLKNAKQLLR KFGVPEEEIK KLSRWEVIDV VRTMSTEQAR SG EGPMSKF ARGSRFSVAE HQERYKEECQ RIFDLQNKVL SSTEVLSTDT DSSSAEDSDF EEMGKNIENM LQNKKTSSQL SRE REEQER KELQRMLLAA GSAASGNNHR DDDTASVTSL NSSATGRCLK IYRTFRDEEG KEYVRCETVR KPAVIDAYVR IRTT KDEEF IRKFALFDEQ HREEMRKERR RIQEQLRRLK RNQEKEKLKG PPEKKPKKMK ERPDLKLKCG ACGAIGHMRT NKFCP LYYQ TNAPPSNPVA MTEEQEEELE KTVIHNDNEE LIKVEGTKIV LGKQLIESAD EVRRKSLVLK FPKQQLPPKK KRRVGT TVH CDYLNRPHKS IHRRRTDPMV TLSSILESII NDMRDLPNTY PFHTPVNAKV VKDYYKIITR PMDLQTLREN VRKRLYP SR EEFREHLELI VKNSATYNGP KHSLTQISQS MLDLCDEKLK EKEDKLARLE KAINPLLDDD DQVAFSFILD NIVTQKMM A VPDSWPFHHP VNKKFVPDYY KVIVNPMDLE TIRKNISKHK YQSRESFLDD VNLILANSVK YNGPESQYTK TAQEIVNVC YQTLTEYDEH LTQLEKDICT AKEAALEEAE LESLDPMTPG PYTPQPPDLY DTNTSLSMSR DASVFQDESN MSVLDIPSAT PEKQVTQEG EDGDGDLADE EEGTVQQPQA SVLYEDLLMS EGEDDEEDAG SDEEGDNPFS AIQLSESGSD SDVGSGGIRP K QPRMLQEN TRMDMENEES MMSYEGDGGE ASHGLEDSNI SYGSYEEPDP KSNTQDTSFS SIGGYEVSEE EEDEEEEEQR SG PSVLSQV HLSEDEEDSE DFHSIAGDSD LDSDE UniProtKB: Transcription initiation factor TFIID subunit 1 |
-Macromolecule #2: Transcription initiation factor TFIID subunit 2
| Macromolecule | Name: Transcription initiation factor TFIID subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 137.159984 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPLTGVEPAR MNRKKGDKGF ESPRPYKLTH QVVCINNINF QRKSVVGFVE LTIFPTVANL NRIKLNSKQC RIYRVRINDL EAAFIYNDP TLEVCHSESK QRNLNYFSNA YAAAVSAVDP DAGNGELCIK VPSELWKHVD ELKVLKIHIN FSLDQPKGGL H FVVPSVEG ...String: MPLTGVEPAR MNRKKGDKGF ESPRPYKLTH QVVCINNINF QRKSVVGFVE LTIFPTVANL NRIKLNSKQC RIYRVRINDL EAAFIYNDP TLEVCHSESK QRNLNYFSNA YAAAVSAVDP DAGNGELCIK VPSELWKHVD ELKVLKIHIN FSLDQPKGGL H FVVPSVEG SMAERGAHVF SCGYQNSTRF WFPCVDSYSE LCTWKLEFTV DAAMVAVSNG DLVETVYTHD MRKKTFHYML TI PTAASNI SLAIGPFEIL VDPYMHEVTH FCLPQLLPLL KHTTSYLHEV FEFYEEILTC RYPYSCFKTV FIDEAYVEVA AYA SMSIFS TNLLHSAMII DETPLTRRCL AQSLAQQFFG CFISRMSWSD EWVLKGISGY IYGLWMKKTF GVNEYRHWIK EELD KIVAY ELKTGGVLLH PIFGGGKEKD NPASHLHFSI KHPHTLSWEY YSMFQCKAHL VMRLIENRIS MEFMLQVFNK LLSLA STAS SQKFQSHMWS QMLVSTSGFL KSISNVSGKD IQPLIKQWVD QSGVVKFYGS FAFNRKRNVL ELEIKQDYTS PGTQKY VGP LKVTVQELDG SFNHTLQIEE NSLKHDIPCH SKSRRNKKKK IPLMNGEEVD MDLSAMDADS PLLWIRIDPD MSVLRKV EF EQADFMWQYQ LRYERDVVAQ QESILALEKF PTPASRLALT DILEQEQCFY RVRMSACFCL AKIANSMVST WTGPPAMK S LFTRMFCCKS CPNIVKTNNF MSFQSYFLQK TMPVAMALLR DVHNLCPKEV LTFILDLIKY NDNRKNKFSD NYYRAEMID ALANSVTPAV SVNNEVRTLD NLNPDVRLIL EEITRFLNME KLLPSYRHTI TVSCLRAIRV LQKNGHVPSD PALFKSYAEY GHFVDIRIA ALEAVVDYTK VDRSYEELQW LLNMIQNDPV PYVRHKILNM LTKNPPFTKN MESPLCNEAL VDQLWKLMNS G TSHDWRLR CGAVDLYFTL FGLSRPSCLP LPELGLVLNL KEKKAVLNPT IIPESVAGNQ EAANNPSSHP QLVGFQNPFS SS QDEEEID MDTVHDSQAF ISHHLNMLER PSTPGLSKYR PASSRSALIP QHSAGCDSTP TTKPQWSLEL ARKGTGKEQA PLE MSMHPA ASAPLSVFTK ESTASKHSDH HHHHHHEHKK KKKKHKHKHK HKHKHDSKEK DKEPFTFSSP ASGRSIRSPS LSD UniProtKB: Transcription initiation factor TFIID subunit 2 |
-Macromolecule #3: Transcription initiation factor TFIID subunit 6
| Macromolecule | Name: Transcription initiation factor TFIID subunit 6 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.749297 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAEEKKLKLS NTVLPSESMK VVAESMGIAQ IQEETCQLLT DEVSYRIKEI AQDALKFMHM GKRQKLTTSD IDYALKLKNV EPLYGFHAQ EFIPFRFASG GGRELYFYEE KEVDLSDIIN TPLPRVPLDV CLKAHWLSIE GCQPAIPENP PPAPKEQQKA E ATEPLKSA ...String: MAEEKKLKLS NTVLPSESMK VVAESMGIAQ IQEETCQLLT DEVSYRIKEI AQDALKFMHM GKRQKLTTSD IDYALKLKNV EPLYGFHAQ EFIPFRFASG GGRELYFYEE KEVDLSDIIN TPLPRVPLDV CLKAHWLSIE GCQPAIPENP PPAPKEQQKA E ATEPLKSA KPGQEEDGPL KGKGQGATTA DGKGKEKKAP PLLEGAPLRL KPRSIHELSV EQQLYYKEIT EACVGSCEAK RA EALQSIA TDPGLYQMLP RFSTFISEGV RVNVVQNNLA LLIYLMRMVK ALMDNPTLYL EKYVHELIPA VMTCIVSRQL CLR PDVDNH WALRDFAARL VAQICKHFST TTNNIQSRIT KTFTKSWVDE KTPWTTRYGS IAGLAELGHD VIKTLILPRL QQEG ERIRS VLDGPVLSNI DRIGADHVQS LLLKHCAPVL AKLRPPPDNQ DAYRAEFGSL GPLLCSQVVK ARAQAALQAQ QVNRT TLTI TQPRPTLTLS QAPQPGPRTP GLLKVPGSIA LPVQTLVSAR AAAPPQPSPP PTKFIVMSSS SSAPSTQQVL SLSTSA PGS GSTTTSPVTT TVPSVQPIVK LVSTATTAPP STAPSGPGSV QKYIVVSLPP TGEGKGGPTS HPSPVPPPAS SPSPLSG SA LCGGKQEAGD SPPPAPGTPK ANGSQPNSGS PQPAP UniProtKB: Transcription initiation factor TFIID subunit 6 |
-Macromolecule #4: Transcription initiation factor TFIID subunit 7
| Macromolecule | Name: Transcription initiation factor TFIID subunit 7 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.325117 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSKSKDDAPH ELESQFILRL PPEYASTVRR AVQSGHVNLK DRLTIELHPD GRHGIVRVDR VPLASKLVDL PCVMESLKTI DKKTFYKTA DICQMLVSTV DGDLYPPVEE PVASTDPKAS KKKDKDKEKK FIWNHGITLP LKNVRKRRFR KTAKKKYIES P DVEKEVKR ...String: MSKSKDDAPH ELESQFILRL PPEYASTVRR AVQSGHVNLK DRLTIELHPD GRHGIVRVDR VPLASKLVDL PCVMESLKTI DKKTFYKTA DICQMLVSTV DGDLYPPVEE PVASTDPKAS KKKDKDKEKK FIWNHGITLP LKNVRKRRFR KTAKKKYIES P DVEKEVKR LLSTDAEAVS TRWEIIAEDE TKEAENQGLD ISSPGMSGHR QGHDSLEHDE LREIFNDLSS SSEDEDETQH QD EEDINII DTEEDLERQL QDKLNESDEQ HQENEGTNQL VMGIQKQIDN MKGKLQETQD RAKRQEDLIM KVENLALKNR FQA VLDELK QKEDREKEQL SSLQEELESL LEK UniProtKB: Transcription initiation factor TFIID subunit 7 |
-Macromolecule #5: Transcription initiation factor TFIID subunit 8
| Macromolecule | Name: Transcription initiation factor TFIID subunit 8 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.304359 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADAAATAGA GGSGTRSGSK QSTNPADNYH LARRRTLQVV VSSLLTEAGF ESAEKASVET LTEMLQSYIS EIGRSAKSYC EHTARTQPT LSDIVVTLVE MGFNVDTLPA YAKRSQRMVI TAPPVTNQPV TPKALTAGQN RPHPPHIPSH FPEFPDPHTY I KTPTYREP ...String: MADAAATAGA GGSGTRSGSK QSTNPADNYH LARRRTLQVV VSSLLTEAGF ESAEKASVET LTEMLQSYIS EIGRSAKSYC EHTARTQPT LSDIVVTLVE MGFNVDTLPA YAKRSQRMVI TAPPVTNQPV TPKALTAGQN RPHPPHIPSH FPEFPDPHTY I KTPTYREP VSDYQVLREK AASQRRDVER ALTRFMAKTG ETQSLFKDDV STFPLIAARP FTIPYLTALL PSELEMQQME ET DSSEQDE QTDTENLALH ISMEDSGAEK ENTSVLQQNP SLSGSRNGEE NIIDNPYLRP VKKPKIRRKK SLS UniProtKB: Transcription initiation factor TFIID subunit 8 |
-Macromolecule #6: DNA (45-MER)
| Macromolecule | Name: DNA (45-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.317238 KDa |
| Sequence | String: (DC)(DT)(DA)(DT)(DA)(DA)(DA)(DA)(DG)(DG) (DG)(DG)(DG)(DT)(DG)(DG)(DG)(DG)(DG)(DC) (DG)(DC)(DG)(DT)(DT)(DC)(DG)(DT)(DC) (DC)(DT)(DC)(DA)(DG)(DT)(DC)(DG)(DC)(DG) (DA) (DT)(DC)(DG)(DA)(DA)(DC) ...String: (DC)(DT)(DA)(DT)(DA)(DA)(DA)(DA)(DG)(DG) (DG)(DG)(DG)(DT)(DG)(DG)(DG)(DG)(DG)(DC) (DG)(DC)(DG)(DT)(DT)(DC)(DG)(DT)(DC) (DC)(DT)(DC)(DA)(DG)(DT)(DC)(DG)(DC)(DG) (DA) (DT)(DC)(DG)(DA)(DA)(DC)(DA)(DC) (DT)(DC)(DG)(DA)(DG)(DC)(DC)(DG)(DA)(DG) (DC)(DA) (DG)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT)(DA)(DC)(DG) |
-Macromolecule #7: DNA (45-MER)
| Macromolecule | Name: DNA (45-MER) / type: dna / ID: 7 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.814336 KDa |
| Sequence | String: (DA)(DT)(DC)(DC)(DA)(DT)(DG)(DG)(DT)(DC) (DC)(DG)(DT)(DA)(DG)(DG)(DC)(DA)(DC)(DG) (DT)(DC)(DT)(DG)(DC)(DT)(DC)(DG)(DG) (DC)(DT)(DC)(DG)(DA)(DG)(DT)(DG)(DT)(DT) (DC) (DG)(DA)(DT)(DC)(DG)(DC) ...String: (DA)(DT)(DC)(DC)(DA)(DT)(DG)(DG)(DT)(DC) (DC)(DG)(DT)(DA)(DG)(DG)(DC)(DA)(DC)(DG) (DT)(DC)(DT)(DG)(DC)(DT)(DC)(DG)(DG) (DC)(DT)(DC)(DG)(DA)(DG)(DT)(DG)(DT)(DT) (DC) (DG)(DA)(DT)(DC)(DG)(DC)(DG)(DA) (DC)(DT)(DG)(DA)(DG)(DG)(DA)(DC)(DG)(DA) (DA)(DC) (DG)(DC)(DG)(DC)(DC)(DC)(DC) (DC)(DA)(DC)(DC)(DC)(DC)(DC)(DT)(DT)(DT) (DT)(DA)(DT) (DA)(DG)(DG)(DC)(DG)(DC) (DC)(DC)(DT)(DT)(DC)(DG)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Grid | Material: GOLD / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.04 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 121285 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7egh: |
 Movie
Movie Controller
Controller















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















