+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31128 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
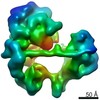
| Title | TFIID-based core PIC on TAF7 promoter | |||||||||
 Map data Map data | TFIID-based core PIC on TAF7 promoter | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of MHC class I biosynthetic process / response to paraquat / spermine transport / SAGA complex assembly / lateral mesodermal cell differentiation / DNA-templated transcription open complex formation / allantois development / pre-snoRNP complex / positive regulation of androgen receptor signaling pathway / TFIIH-class transcription factor complex binding ...negative regulation of MHC class I biosynthetic process / response to paraquat / spermine transport / SAGA complex assembly / lateral mesodermal cell differentiation / DNA-templated transcription open complex formation / allantois development / pre-snoRNP complex / positive regulation of androgen receptor signaling pathway / TFIIH-class transcription factor complex binding / negative regulation of protein autoubiquitination / positive regulation of core promoter binding / negative regulation of MHC class II biosynthetic process / transcription factor TFTC complex / RNA polymerase II core complex assembly / regulation of cell cycle G1/S phase transition / RNA polymerase I general transcription initiation factor activity / RNA polymerase transcription factor SL1 complex / meiotic sister chromatid cohesion / phosphatase activator activity / SLIK (SAGA-like) complex / RNA polymerase III general transcription initiation factor activity / TFIIF-class transcription factor complex binding / transcriptional start site selection at RNA polymerase II promoter / RNA polymerase I core promoter sequence-specific DNA binding / hepatocyte differentiation / transcription factor TFIIF complex / positive regulation of response to cytokine stimulus / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Transcription Initiation From Type 3 Promoter / Formation of RNA Pol II elongation complex / Formation of the Early Elongation Complex / Transcriptional regulation by small RNAs / RNA Polymerase II Pre-transcription Events / TP53 Regulates Transcription of DNA Repair Genes / FGFR2 alternative splicing / RNA polymerase II transcribes snRNA genes / mRNA Capping / mRNA Splicing - Minor Pathway / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Elongation / RNA Polymerase II Transcription Initiation And Promoter Clearance / RNA Pol II CTD phosphorylation and interaction with CE / Estrogen-dependent gene expression / Formation of TC-NER Pre-Incision Complex / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / mRNA Splicing - Major Pathway / transcription factor TFIIA complex / female germ cell nucleus / C2H2 zinc finger domain binding / maintenance of protein location in nucleus / RNA Polymerase III Abortive And Retractive Initiation / male pronucleus / histone H3K4me3 reader activity / female pronucleus / host-mediated activation of viral transcription / germinal vesicle / RNA polymerase II general transcription initiation factor binding / RNA polymerase binding / nuclear vitamin D receptor binding / regulation of fat cell differentiation / nuclear thyroid hormone receptor binding / box C/D snoRNP assembly / SAGA complex / Abortive elongation of HIV-1 transcript in the absence of Tat / FGFR2 alternative splicing / transcription preinitiation complex / RNA Polymerase I Transcription Termination / Viral Messenger RNA Synthesis / Signaling by FGFR2 IIIa TM / inner cell mass cell proliferation / protein acetylation / negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / RNA polymerase II general transcription initiation factor activity / cell division site / transcription factor TFIID complex / limb development / histone acetyltransferase binding / acetyltransferase activity / RNA Pol II CTD phosphorylation and interaction with CE during HIV infection / RNA Pol II CTD phosphorylation and interaction with CE / midbrain development / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / cellular response to ATP / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / regulation of RNA splicing / mRNA Splicing - Minor Pathway Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 19.3 Å | |||||||||
 Authors Authors | Chen X / Yilun Q / Hou H / Wang X / Wu Z / Li J / Xu Y | |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Structural insights into preinitiation complex assembly on core promoters. Authors: Xizi Chen / Yilun Qi / Zihan Wu / Xinxin Wang / Jiabei Li / Dan Zhao / Haifeng Hou / Yan Li / Zishuo Yu / Weida Liu / Mo Wang / Yulei Ren / Ze Li / Huirong Yang / Yanhui Xu /  Abstract: Transcription factor IID (TFIID) recognizes core promoters and supports preinitiation complex (PIC) assembly for RNA polymerase II (Pol II)-mediated eukaryotic transcription. We determined the ...Transcription factor IID (TFIID) recognizes core promoters and supports preinitiation complex (PIC) assembly for RNA polymerase II (Pol II)-mediated eukaryotic transcription. We determined the structures of human TFIID-based PIC in three stepwise assembly states and revealed two-track PIC assembly: stepwise promoter deposition to Pol II and extensive modular reorganization on track I (on TATA-TFIID-binding element promoters) versus direct promoter deposition on track II (on TATA-only and TATA-less promoters). The two tracks converge at an ~50-subunit holo PIC in identical conformation, whereby TFIID stabilizes PIC organization and supports loading of cyclin-dependent kinase (CDK)-activating kinase (CAK) onto Pol II and CAK-mediated phosphorylation of the Pol II carboxyl-terminal domain. Unexpectedly, TBP of TFIID similarly bends TATA box and TATA-less promoters in PIC. Our study provides structural visualization of stepwise PIC assembly on highly diversified promoters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31128.map.gz emd_31128.map.gz | 25.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31128-v30.xml emd-31128-v30.xml emd-31128.xml emd-31128.xml | 7.7 KB 7.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31128.png emd_31128.png | 35.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31128 http://ftp.pdbj.org/pub/emdb/structures/EMD-31128 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31128 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31128 | HTTPS FTP |
-Related structure data
| Related structure data |  7edxC  7eg7C  7eg8C  7eg9C  7egaC  7egbC  7egcC  7egdC  7egeC  7egfC  7eggC  7eghC  7egiC  7egjC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31128.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31128.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TFIID-based core PIC on TAF7 promoter | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.284 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TFIID-based core PIC on TAF7 promoter
| Entire | Name: TFIID-based core PIC on TAF7 promoter |
|---|---|
| Components |
|
-Supramolecule #1: TFIID-based core PIC on TAF7 promoter
| Supramolecule | Name: TFIID-based core PIC on TAF7 promoter / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 19.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 4753 |
|---|---|
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















