[English] 日本語
 Yorodumi
Yorodumi- EMDB-2283: Human TFIID Binds to Core Promoter DNA in a Reorganized Structura... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2283 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human TFIID Binds to Core Promoter DNA in a Reorganized Structural State | |||||||||
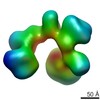
 Map data Map data | 3D reconstruction of canonical conformation of human TFIID in the presence of TFIIA and super core promoter DNA at 32A | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TFIID / transcription / cryo-electron microscopy / TFIIA / core promoter / RNA polymerase II | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 32.0 Å | |||||||||
 Authors Authors | Cianfrocco MA / Kassavetis GA / Grob P / Fang J / Juven-Gershon T / Kadonaga JT / Nogales E | |||||||||
 Citation Citation |  Journal: Cell / Year: 2013 Journal: Cell / Year: 2013Title: Human TFIID binds to core promoter DNA in a reorganized structural state. Authors: Michael A Cianfrocco / George A Kassavetis / Patricia Grob / Jie Fang / Tamar Juven-Gershon / James T Kadonaga / Eva Nogales /  Abstract: A mechanistic description of metazoan transcription is essential for understanding the molecular processes that govern cellular decisions. To provide structural insights into the DNA recognition step ...A mechanistic description of metazoan transcription is essential for understanding the molecular processes that govern cellular decisions. To provide structural insights into the DNA recognition step of transcription initiation, we used single-particle electron microscopy (EM) to visualize human TFIID with promoter DNA. This analysis revealed that TFIID coexists in two predominant and distinct structural states that differ by a 100 Å translocation of TFIID's lobe A. The transition between these structural states is modulated by TFIIA, as the presence of TFIIA and promoter DNA facilitates the formation of a rearranged state of TFIID that enables promoter recognition and binding. DNA labeling and footprinting, together with cryo-EM studies, were used to map the locations of TATA, Initiator (Inr), motif ten element (MTE), and downstream core promoter element (DPE) promoter motifs within the TFIID-TFIIA-DNA structure. The existence of two structurally and functionally distinct forms of TFIID suggests that the different conformers may serve as specific targets for the action of regulatory factors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2283.map.gz emd_2283.map.gz | 3.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2283-v30.xml emd-2283-v30.xml emd-2283.xml emd-2283.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2283.png EMD-2283.png | 46.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2283 http://ftp.pdbj.org/pub/emdb/structures/EMD-2283 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2283 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2283 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2283.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2283.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of canonical conformation of human TFIID in the presence of TFIIA and super core promoter DNA at 32A | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.48 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TFIID-TFIIA-SCP (canonical)
| Entire | Name: TFIID-TFIIA-SCP (canonical) |
|---|---|
| Components |
|
-Supramolecule #1000: TFIID-TFIIA-SCP (canonical)
| Supramolecule | Name: TFIID-TFIIA-SCP (canonical) / type: sample / ID: 1000 / Number unique components: 3 |
|---|---|
| Molecular weight | Experimental: 1.4 MDa / Theoretical: 1.4 MDa |
-Macromolecule #1: Transcription factor II-D
| Macromolecule | Name: Transcription factor II-D / type: protein_or_peptide / ID: 1 / Name.synonym: TFIID / Number of copies: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: HeLa / synonym: Human / Organelle: Nucleus Homo sapiens (human) / Strain: HeLa / synonym: Human / Organelle: Nucleus |
| Molecular weight | Experimental: 1.4 MDa / Theoretical: 1.4 MDa |
-Macromolecule #2: Transcription factor II-A
| Macromolecule | Name: Transcription factor II-A / type: protein_or_peptide / ID: 2 / Name.synonym: TFIIA / Number of copies: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: HeLa / synonym: Human / Organelle: Nucleus Homo sapiens (human) / Strain: HeLa / synonym: Human / Organelle: Nucleus |
| Molecular weight | Experimental: 1.4 MDa / Theoretical: 1.4 MDa |
-Macromolecule #3: Transcription factor SCP
| Macromolecule | Name: Transcription factor SCP / type: protein_or_peptide / ID: 3 / Name.synonym: SCP / Number of copies: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: HeLa / synonym: Human / Organelle: Nucleus Homo sapiens (human) / Strain: HeLa / synonym: Human / Organelle: Nucleus |
| Molecular weight | Experimental: 1.4 MDa / Theoretical: 1.4 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.8 Details: 20 mM HEPES pH 7.8, 0.2 mM EDTA, 4 mM MgCl2, ~1% glycerol, 1 mM DTT, 3% trehalose, 50 mM KCl, 0.05% NP-40, |
| Grid | Details: 400 mesh copper grid on C-flat support carbon with a thin carbon support over 4 um holes space 2 um apart. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK II Method: Sample was incubated on grid for 30 seconds before blotting 6.5s at -1 offset. The grid was washed in 4 ul buffer to blot again for 6.5 s at -1 offset prior to plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective and condenser lens astigmatism was corrected at 80,000 times magnification |
| Date | May 10, 2011 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 1037 / Average electron dose: 25 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 10500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 80000 |
| Sample stage | Specimen holder: Gatan 626 liquid nitrogen cooled / Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Particles were prepared within the APPION processing environment. The particles were automatically selected using Signature and extracted from phase flipped micrographs using values calculated by CTFFIND3. After projection matching within EMAN2/Sparx, the final map was further refined in FREALIGN. |
|---|---|
| CTF correction | Details: Per micrograph for phase flipping, per particles for refinement in FREALIGN |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 32.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: Sparx, EMAN2, FREALIGN / Number images used: 8715 |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















