[English] 日本語
 Yorodumi
Yorodumi- EMDB-22336: Cryo-EM structure of ATP-bound fully inactive AMPK in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22336 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody | |||||||||
 Map data Map data | Structure of AMPK in complex with CpdC and Fab-nanobody | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AMPK / ATP / fully inactive / KD-displaced / TRANSFERASE-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of glucosylceramide biosynthetic process / [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase / [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity / regulation of stress granule assembly / positive regulation of mitochondrial transcription / negative regulation of tubulin deacetylation / AMPK inhibits chREBP transcriptional activation activity / histone H2BS36 kinase activity / cold acclimation / cAMP-dependent protein kinase regulator activity ...negative regulation of glucosylceramide biosynthetic process / [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase / [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity / regulation of stress granule assembly / positive regulation of mitochondrial transcription / negative regulation of tubulin deacetylation / AMPK inhibits chREBP transcriptional activation activity / histone H2BS36 kinase activity / cold acclimation / cAMP-dependent protein kinase regulator activity / AMP-activated protein kinase activity / lipid droplet disassembly / Lipophagy / regulation of carbon utilization / positive regulation of skeletal muscle tissue development / CAMKK-AMPK signaling cascade / import into nucleus / regulation of vesicle-mediated transport / nucleotide-activated protein kinase complex / negative regulation of hepatocyte apoptotic process / Energy dependent regulation of mTOR by LKB1-AMPK / Carnitine shuttle / positive regulation of T cell mediated immune response to tumor cell / tau-protein kinase / protein kinase regulator activity / negative regulation of TOR signaling / Activation of PPARGC1A (PGC-1alpha) by phosphorylation / response to caffeine / regulation of glycolytic process / : / cAMP-dependent protein kinase activity / tau-protein kinase activity / protein localization to lipid droplet / AMP binding / cellular response to stress / Macroautophagy / cholesterol biosynthetic process / lipid biosynthetic process / fatty acid oxidation / motor behavior / positive regulation of protein kinase activity / cellular response to ethanol / carbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / fatty acid homeostasis / negative regulation of lipid catabolic process / response to UV / cellular response to glucose starvation / energy homeostasis / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / Activation of AMPK downstream of NMDARs / positive regulation of protein localization / positive regulation of adipose tissue development / negative regulation of TORC1 signaling / cellular response to nutrient levels / positive regulation of autophagy / positive regulation of gluconeogenesis / negative regulation of insulin receptor signaling pathway / cellular response to calcium ion / regulation of microtubule cytoskeleton organization / positive regulation of glycolytic process / response to activity / response to gamma radiation / TP53 Regulates Metabolic Genes / Translocation of SLC2A4 (GLUT4) to the plasma membrane / cellular response to glucose stimulus / positive regulation of cholesterol biosynthetic process / neuron cellular homeostasis / ADP binding / regulation of circadian rhythm / autophagy / response to estrogen / cellular response to xenobiotic stimulus / tau protein binding / glucose metabolic process / Wnt signaling pathway / cellular response to hydrogen peroxide / positive regulation of T cell activation / fatty acid biosynthetic process / rhythmic process / glucose homeostasis / cellular response to prostaglandin E stimulus / positive regulation of cold-induced thermogenesis / outer membrane-bounded periplasmic space / cellular response to oxidative stress / spermatogenesis / cellular response to hypoxia / Regulation of TP53 Activity through Phosphorylation / response to hypoxia / protein phosphorylation / protein kinase activity / non-specific serine/threonine protein kinase / apical plasma membrane / regulation of cell cycle / cilium / nuclear speck / ciliary basal body / negative regulation of gene expression Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) / Homo sapiens (human) / synthetic construct (others) /  | |||||||||
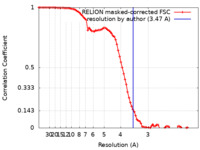
| Method | single particle reconstruction / cryo EM / Resolution: 3.47 Å | |||||||||
 Authors Authors | Yan Y / Murkherjee S / Zhou XE / Xu TH / Xu HE / Kossiakoff AA / Melcher K | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Structure of an AMPK complex in an inactive, ATP-bound state. Authors: Yan Yan / Somnath Mukherjee / Kaleeckal G Harikumar / Timothy S Strutzenberg / X Edward Zhou / Kelly Suino-Powell / Ting-Hai Xu / Ryan D Sheldon / Jared Lamp / Joseph S Brunzelle / Katarzyna ...Authors: Yan Yan / Somnath Mukherjee / Kaleeckal G Harikumar / Timothy S Strutzenberg / X Edward Zhou / Kelly Suino-Powell / Ting-Hai Xu / Ryan D Sheldon / Jared Lamp / Joseph S Brunzelle / Katarzyna Radziwon / Abigail Ellis / Scott J Novick / Irving E Vega / Russell G Jones / Laurence J Miller / H Eric Xu / Patrick R Griffin / Anthony A Kossiakoff / Karsten Melcher /   Abstract: Adenosine monophosphate (AMP)-activated protein kinase (AMPK) regulates metabolism in response to the cellular energy states. Under energy stress, AMP stabilizes the active AMPK conformation, in ...Adenosine monophosphate (AMP)-activated protein kinase (AMPK) regulates metabolism in response to the cellular energy states. Under energy stress, AMP stabilizes the active AMPK conformation, in which the kinase activation loop (AL) is protected from protein phosphatases, thus keeping the AL in its active, phosphorylated state. At low AMP:ATP (adenosine triphosphate) ratios, ATP inhibits AMPK by increasing AL dynamics and accessibility. We developed conformation-specific antibodies to trap ATP-bound AMPK in a fully inactive, dynamic state and determined its structure at 3.5-angstrom resolution using cryo-electron microscopy. A 180° rotation and 100-angstrom displacement of the kinase domain fully exposes the AL. On the basis of the structure and supporting biophysical data, we propose a multistep mechanism explaining how adenine nucleotides and pharmacological agonists modulate AMPK activity by altering AL phosphorylation and accessibility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22336.map.gz emd_22336.map.gz | 3.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22336-v30.xml emd-22336-v30.xml emd-22336.xml emd-22336.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22336_fsc.xml emd_22336_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_22336.png emd_22336.png | 182.1 KB | ||
| Filedesc metadata |  emd-22336.cif.gz emd-22336.cif.gz | 7.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22336 http://ftp.pdbj.org/pub/emdb/structures/EMD-22336 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22336 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22336 | HTTPS FTP |
-Related structure data
| Related structure data |  7jhgMC  7jhhC  7jijC  7m74C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22336.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22336.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of AMPK in complex with CpdC and Fab-nanobody | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.029 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Cryo-EM structure of ATP-bound fully inactive AMPK in complex wit...
+Supramolecule #1: Cryo-EM structure of ATP-bound fully inactive AMPK in complex wit...
+Supramolecule #2: ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compo...
+Supramolecule #3: Fab-nanobody
+Macromolecule #1: 5'-AMP-activated protein kinase catalytic subunit alpha-1
+Macromolecule #2: 5'-AMP-activated protein kinase subunit beta-2
+Macromolecule #3: 5'-AMP-activated protein kinase subunit gamma-1
+Macromolecule #4: Maltodextrin-binding protein
+Macromolecule #5: Fab light chain
+Macromolecule #6: Fab heavy chain
+Macromolecule #7: Nanobody
+Macromolecule #9: 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a...
+Macromolecule #10: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #11: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #12: ADENOSINE MONOPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 6157 / Average exposure time: 0.2 sec. / Average electron dose: 83.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Output model |  PDB-7jhg: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)