-Search query
-Search result
Showing 1 - 50 of 1,565 items for (author: ye & rd)

EMDB entry, No image
EMDB-16860: 
Ivabradine bound to HCN4 channel

PDB-8ofi: 
Ivabradine bound to HCN4 channel

EMDB-43139: 
SARS-CoV-2 Spike S2 bound to Fab 54043-5

EMDB-18990: 
CryoEM map of tau PHF sarkosyl-extracted from a human AD patient (associated with in situ tomography)

EMDB-43011: 
Phosphorylated, ATP-bound, E1371Q human cystic fibrosis transmembrane conductance regulator (E1371Q-CFTR)

EMDB-43014: 
Phosphorylated, ATP-bound, inhibitor 172-bound E1371Q human cystic fibrosis transmembrane conductance regulator
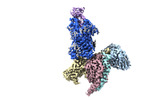
EMDB-36869: 
Cryo-EM structure of CXCR4 in complex with CXCL12

EMDB-50148: 
Tau PHF subtomogram average relating to CS1 extended data Figure 9A

EMDB-50152: 
Tau PHF subtomogram average relating to CS2 Figure 3i-j.

EMDB-50153: 
Tau PHF subtomogram average relating to CS3 extended data Figure 9c

EMDB-50155: 
Tau PHF subtomogram average relating to CS4 extended data Figure 9d

EMDB-50156: 
Tau PHF subtomogram average relating to CS5 extended data Figure 9b

EMDB-50157: 
Tau PHF subtomogram average relating to CS6 extended data Figure 9e

EMDB-50159: 
Tau PHF subtomogram average relating to CS7 extended data Figure 9f

EMDB-50160: 
Tau PHF subtomogram average relating to LOL1_PHF Figure 4g-h

EMDB-50161: 
Tau SF subtomogram average relating to LOL1_SF Figure 4g-h

EMDB-50162: 
Tau SF subtomogram average relating to LOL2_SF Figure 4i-j

EMDB-50580: 
SOLIST cryo-tomogram of native left ventricle mouse heart muscle #1

EMDB-50582: 
SOLIST native mouse heart muscle tomogram #2

EMDB-41433: 
Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter

EMDB-41437: 
Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter

EMDB-41439: 
Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter

EMDB-41448: 
Escherichia coli RNA polymerase unwinding intermediate (I1c) at the lambda PR promoter

EMDB-41456: 
Escherichia coli RNA polymerase closed complex intermediate at the lambda PR promoter

PDB-8to1: 
Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter

PDB-8to6: 
Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter

PDB-8to8: 
Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter

PDB-8toe: 
Escherichia coli RNA polymerase unwinding intermediate (I1c) at the lambda PR promoter

PDB-8tom: 
Escherichia coli RNA polymerase closed complex intermediate at the lambda PR promoter

EMDB-19066: 
TREK2 in OGNG/CHS detergent micelle with biparatopic inhibitory nanobody Nb6158

EMDB-40825: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10

PDB-8sx3: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10
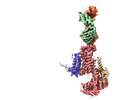
EMDB-37159: 
Cryo-EM structure of NADPH oxidase 2 in complex with p22phox and EROS
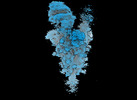
EMDB-18639: 
Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2)

EMDB-18649: 
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab

EMDB-19002: 
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein

PDB-8qsq: 
Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2)

PDB-8qtd: 
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab

PDB-8r8k: 
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein

EMDB-44496: 
Cryo-EM co-structure of AcrB with the CU032 efflux pump inhibitor

EMDB-44500: 
Cryo-EM co-structure of AcrB with the EPM35 efflux pump inhibitor

EMDB-44501: 
Cryo-EM co-structure of AcrB with the CU232 efflux pump inhibitor
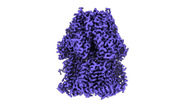
EMDB-44506: 
Cryo-EM co-structure of AcrB with CU244

PDB-9bfh: 
Cryo-EM co-structure of AcrB with the CU032 efflux pump inhibitor

PDB-9bfm: 
Cryo-EM co-structure of AcrB with the EPM35 efflux pump inhibitor

PDB-9bfn: 
Cryo-EM co-structure of AcrB with the CU232 efflux pump inhibitor

PDB-9bft: 
Cryo-EM co-structure of AcrB with CU244

EMDB-40674: 
Subtomogram average of immature PhiKZ Major Capsid Protein from tubular arrays

EMDB-17189: 
Structure of the human neutral amino acid transporter ASCT2 in complex with nanobody 469

EMDB-17192: 
Complex of human ASCT2 with Syncytin-1
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
