[English] 日本語
 Yorodumi
Yorodumi- EMDB-40674: Subtomogram average of immature PhiKZ Major Capsid Protein from t... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of immature PhiKZ Major Capsid Protein from tubular arrays | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | major capsid protein / HK97-fold / VIRAL PROTEIN | |||||||||||||||
| Function / homology | viral capsid / Major capsid protein Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Phikzvirus phiKZ Phikzvirus phiKZ | |||||||||||||||
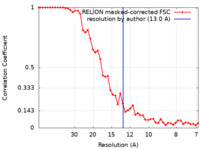
| Method | subtomogram averaging / cryo EM / Resolution: 13.0 Å | |||||||||||||||
 Authors Authors | Laughlin TG / Villa E | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: An intron endonuclease facilitates interference competition between coinfecting viruses. Authors: Erica A Birkholz / Chase J Morgan / Thomas G Laughlin / Rebecca K Lau / Amy Prichard / Sahana Rangarajan / Gabrielle N Meza / Jina Lee / Emily Armbruster / Sergey Suslov / Kit Pogliano / ...Authors: Erica A Birkholz / Chase J Morgan / Thomas G Laughlin / Rebecca K Lau / Amy Prichard / Sahana Rangarajan / Gabrielle N Meza / Jina Lee / Emily Armbruster / Sergey Suslov / Kit Pogliano / Justin R Meyer / Elizabeth Villa / Kevin D Corbett / Joe Pogliano /  Abstract: Introns containing homing endonucleases are widespread in nature and have long been assumed to be selfish elements that provide no benefit to the host organism. These genetic elements are common in ...Introns containing homing endonucleases are widespread in nature and have long been assumed to be selfish elements that provide no benefit to the host organism. These genetic elements are common in viruses, but whether they confer a selective advantage is unclear. In this work, we studied intron-encoded homing endonuclease gp210 in bacteriophage ΦPA3 and found that it contributes to viral competition by interfering with the replication of a coinfecting phage, ΦKZ. We show that gp210 targets a specific sequence in ΦKZ, which prevents the assembly of progeny viruses. This work demonstrates how a homing endonuclease can be deployed in interference competition among viruses and provide a relative fitness advantage. Given the ubiquity of homing endonucleases, this selective advantage likely has widespread evolutionary implications in diverse plasmid and viral competition as well as virus-host interactions. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40674.map.gz emd_40674.map.gz | 5.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40674-v30.xml emd-40674-v30.xml emd-40674.xml emd-40674.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40674_fsc.xml emd_40674_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_40674.png emd_40674.png | 55.7 KB | ||
| Masks |  emd_40674_msk_1.map emd_40674_msk_1.map | 8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-40674.cif.gz emd-40674.cif.gz | 5.6 KB | ||
| Others |  emd_40674_half_map_1.map.gz emd_40674_half_map_1.map.gz emd_40674_half_map_2.map.gz emd_40674_half_map_2.map.gz | 5.4 MB 5.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40674 http://ftp.pdbj.org/pub/emdb/structures/EMD-40674 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40674 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40674 | HTTPS FTP |
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40674.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40674.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.457 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40674_msk_1.map emd_40674_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_40674_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_40674_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Immature PhiKZ major capsid protein in a tubular array
| Entire | Name: Immature PhiKZ major capsid protein in a tubular array |
|---|---|
| Components |
|
-Supramolecule #1: Immature PhiKZ major capsid protein in a tubular array
| Supramolecule | Name: Immature PhiKZ major capsid protein in a tubular array type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Phikzvirus phiKZ Phikzvirus phiKZ |
-Macromolecule #1: PhiKZ major capsid protein
| Macromolecule | Name: PhiKZ major capsid protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Phikzvirus phiKZ Phikzvirus phiKZ |
| Sequence | String: MSVHALRELF KHKGEKNYEV FSMEDFIGRL ESEIGLNDSV VSQGRSLISS ISHENFGTVQ ATDIQDAAAI YNKMQMLVND YGFERVSSSD PQVRAREERV RENQITAATM AAIACADETK YIRALRGITK AKASNEDHVK VVQHQFNGPA GGIQVFENGV GLENYNEKSQ ...String: MSVHALRELF KHKGEKNYEV FSMEDFIGRL ESEIGLNDSV VSQGRSLISS ISHENFGTVQ ATDIQDAAAI YNKMQMLVND YGFERVSSSD PQVRAREERV RENQITAATM AAIACADETK YIRALRGITK AKASNEDHVK VVQHQFNGPA GGIQVFENGV GLENYNEKSQ RDFRVVTIGY NLAASRQDEF AERIYPTTVI NPIEGGVVQV LPYIAVMKDV YHEVSGVKMD NEEVNMVEAY RDPSILDDES IALIPALDPA GSNADFFVDP ALVPPYTIKN EQNLTITTAP LKANVRLDLM GNSNANLLIQ RGMLEVSDTI DPAGRLKNLF VLLGGKVVKF KVDRLPRAVF QPDLVGDTRN AVIRFDSDDL VVSGDTTFID GSADGVINDL KTAKLSLRLS VGFGGTISLS KGDSKFGATD TYVDKVLNED GQVMDNADPA VKAILDQLTD LAVIGFELDT RFTNTNRRQR GHLLQTRALQ FRHPIPMHAP VTLPMDTMTD EGPGEVVKAL TVNTNIRNSN NAVKRMLNYL AQLREVVHNG YNRPKFGIIE GALSAVMRPT YRYKELDLEK VIDTIKSKDR WDDVCAAILN CVKAELFPAH RDSNIEAAFR VISGNQDETP MYLFCSDKEI ANYLMTKGDD RTLGAYLKYD IVSTNNQLFD GKLVVIPTRA VQQENDILSW GQFFYVSTVI ADLPITRGGH QVTREIAAIP FNLHVNNIPF ALEFKITGFQ KVMGETQFNG KLADLKP UniProtKB: Major capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: HOMEMADE PLUNGER |
| Details | P. aeruginosa cells expressing PhiPA3 gp210-GFPmut1 infected with PhiKZ |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 1 / Average electron dose: 2.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 7.0 µm / Nominal defocus min: 4.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)