+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab | |||||||||
 Map data Map data | Sharpened map of BA.2.86 spike with XBB7 fab local refinement. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Viral protein/immune system / SARS-CoV-2 / XBB-7 / RBD / Spike / BA.2.86 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvirion component / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...virion component / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Ren J / Duyvesteyn HME / Stuart DI | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep Med / Year: 2024 Journal: Cell Rep Med / Year: 2024Title: A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity. Authors: Chang Liu / Daming Zhou / Aiste Dijokaite-Guraliuc / Piyada Supasa / Helen M E Duyvesteyn / Helen M Ginn / Muneeswaran Selvaraj / Alexander J Mentzer / Raksha Das / Thushan I de Silva / ...Authors: Chang Liu / Daming Zhou / Aiste Dijokaite-Guraliuc / Piyada Supasa / Helen M E Duyvesteyn / Helen M Ginn / Muneeswaran Selvaraj / Alexander J Mentzer / Raksha Das / Thushan I de Silva / Thomas G Ritter / Megan Plowright / Thomas A H Newman / Lizzie Stafford / Barbara Kronsteiner / Nigel Temperton / Yuan Lui / Martin Fellermeyer / Philip Goulder / Paul Klenerman / Susanna J Dunachie / Michael I Barton / Mikhail A Kutuzov / Omer Dushek / / Elizabeth E Fry / Juthathip Mongkolsapaya / Jingshan Ren / David I Stuart / Gavin R Screaton /    Abstract: BA.2.86, a recently described sublineage of SARS-CoV-2 Omicron, contains many mutations in the spike gene. It appears to have originated from BA.2 and is distinct from the XBB variants responsible ...BA.2.86, a recently described sublineage of SARS-CoV-2 Omicron, contains many mutations in the spike gene. It appears to have originated from BA.2 and is distinct from the XBB variants responsible for many infections in 2023. The global spread and plethora of mutations in BA.2.86 has caused concern that it may possess greater immune-evasive potential, leading to a new wave of infection. Here, we examine the ability of BA.2.86 to evade the antibody response to infection using a panel of vaccinated or naturally infected sera and find that it shows marginally less immune evasion than XBB.1.5. We locate BA.2.86 in the antigenic landscape of recent variants and look at its ability to escape panels of potent monoclonal antibodies generated against contemporary SARS-CoV-2 infections. We demonstrate, and provide a structural explanation for, increased affinity of BA.2.86 to ACE2, which may increase transmissibility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18649.map.gz emd_18649.map.gz | 326 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18649-v30.xml emd-18649-v30.xml emd-18649.xml emd-18649.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18649.png emd_18649.png | 51.8 KB | ||
| Filedesc metadata |  emd-18649.cif.gz emd-18649.cif.gz | 7.6 KB | ||
| Others |  emd_18649_half_map_1.map.gz emd_18649_half_map_1.map.gz emd_18649_half_map_2.map.gz emd_18649_half_map_2.map.gz | 322.2 MB 311 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18649 http://ftp.pdbj.org/pub/emdb/structures/EMD-18649 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18649 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18649 | HTTPS FTP |
-Related structure data
| Related structure data |  8qtdMC  8qrfC  8qrgC  8qsqC  8r80C  8r8kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18649.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18649.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of BA.2.86 spike with XBB7 fab local refinement. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7303 Å | ||||||||||||||||||||||||||||||||||||
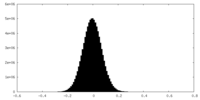
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map of BA.2.86 spike with XBB7 fab local refinement.
| File | emd_18649_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of BA.2.86 spike with XBB7 fab local refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
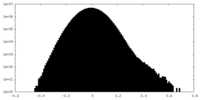
| Density Histograms |
-Half map: #1
| File | emd_18649_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
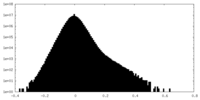
| Density Histograms |
- Sample components
Sample components
-Entire : BA-2.86 variant SARS-CoV2 S protein complexed with XBB-7 fab.
| Entire | Name: BA-2.86 variant SARS-CoV2 S protein complexed with XBB-7 fab. |
|---|---|
| Components |
|
-Supramolecule #1: BA-2.86 variant SARS-CoV2 S protein complexed with XBB-7 fab.
| Supramolecule | Name: BA-2.86 variant SARS-CoV2 S protein complexed with XBB-7 fab. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Spike protein recombinantly expressed using sequence of human-derived BA-2.86 variant of SARS-CoV2. XBB7 fab recombinantely expressed from sequenced fab derived from convalescent sera. |
|---|
-Supramolecule #2: XBB-7 fab
| Supramolecule | Name: XBB-7 fab / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2-#3 Details: XBB-7 fab recombinantely expressed from sequenced fab derived from convalescent sera. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: BA.2.86 variant SARS-CoV2 S protein
| Supramolecule | Name: BA.2.86 variant SARS-CoV2 S protein / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 Details: Spike protein recombinantly expressed using sequence of human-derived BA-2.86 variant of SARS-CoV2. |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein,Fibritin
| Macromolecule | Name: Spike glycoprotein,Fibritin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 142.385828 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVMPLF NLITTTQSYT NSFTRGVYYP DKVFRSSVLH LTQDLFLPFF SNVTWFHAIS GTNGTKRFDN PVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVFIKV CEFQFCNDPF LDVYHKNNKS WMESESGVYS S ANNCTFEY ...String: MFVFLVLLPL VSSQCVMPLF NLITTTQSYT NSFTRGVYYP DKVFRSSVLH LTQDLFLPFF SNVTWFHAIS GTNGTKRFDN PVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVFIKV CEFQFCNDPF LDVYHKNNKS WMESESGVYS S ANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI IGRDFPQGFS ALEPLVDLPI GINITRFQTL LA LNRSYLT PGDSSSGWTA GAADYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPT ESIVRF PNVTNLCPFH EVFNATRFAS VYAWNRTRIS NCVADYSVLY NFAPFFAFKC YGVSPTKLND LCFTNVYADS FVIK GNEVS QIAPGQTGNI ADYNYKLPDD FTGCVIAWNS NKLDSKHSGN YDYWYRLFRK SKLKPFERDI STEIYQAGNK PCKGK GPNC YFPLQSYGFR PTYGVGHQPY RVVVLSFELL HAPATVCGPK KSTNLVKNKC VNFNFNGLTG TGVLTKSNKK FLPFQQ FGR DIVDTTDAVR DPQTLEILDI TPCSFGGVSV ITPGTNTSNQ VAVLYQGVNC TEVSVAIHAD QLTPTWRVYS TGSNVFQ TR AGCLIGAEYV NNSYECDIPI GAGVCASYQT QTKSHGSASS VASQSIIAYT MSLGAENSVA YSNNSIAIPT NFTISVTT E ILPVSMTKTS VDCTMYICGD STECSNLLLQ YGSFCTQLKR ALTGIAVEQD KNTQEVFAQV KQIYKTPPIK YFGGFNFSQ ILPDPSKPSK RSFIEDLLFN KVTLADAGFI KQYGDCLGDI AARDLICAQK FNGLTVLPPL LTDEMIAQYT SALLAGTITS GWTFGAGAA LQIPFAMQMA YRFNGIGVTQ NVLYENQKLI ANQFNSAIGK IQDSLFSTAS ALGKLQDVVN HNAQALNTLV K QLSSKFGA ISSVLNDILS RLDPPEAEVQ IDRLITGRLQ SLQTYVTQQL IRAAEIRASA NLAATKMSEC VLGQSKRVDF CG KGYHLMS FPQSAPHGVV FLHVTYVPAQ EKNFTTAPAI CHDGKAHFPR EGVFVSNGTH WFVTQRNFYE PQIITTDNTF VSG NCDVVI GIVNNTVYDP LQLELDSFKE ELDKYFKNHT SPDVDLGDIS GINASVVNIQ KEIDRLNEVA KNLNESLIDL QELG KYEQG SGYIPEAPRD GQAYVRKDGE WVLLSTFLGR SLEVLFQGPG HHHHHHHHGS AWSHPQFEKG GGSGGGSGGS AWSHP QFEK UniProtKB: Spike glycoprotein, Fibritin |
-Macromolecule #2: XBB-7 fab heavy chain
| Macromolecule | Name: XBB-7 fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.613259 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVESGGG LVQPGGSLRL SCAASGFIFR SFSMSWVRQA PGKGLEWVAN INEDGGEKYY VDSVKGRFTI SRDYAKDSVF LQMNSLRAE DTAVYYCARV GPYYYDSAGY YRRHYHFGMD VWGQGTTVTV SS |
-Macromolecule #3: XBB-7 fab light chain
| Macromolecule | Name: XBB-7 fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.388351 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSALTQPASV SGSPGQSITI SCTGTSSDIG DYNYVSWYQQ HPGKAPKLMI LSNRFSGSKS GNTASLTISG LQAEDEADYY CSSYTGSVT VFGGGTKLTV L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. |
| Vitrification | Cryogen name: ETHANE |
| Details | BA-2.86 Spike with XBB-7 fab in 6-fold excess of S protein. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 9664 / Average electron dose: 50.0 e/Å2 / Details: Images were collected in EER format. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)