+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of CXCR4 in complex with CXCL12 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Chemokine receptor / CXCR4 / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationtelencephalon cell migration / chemokine (C-X-C motif) ligand 12 signaling pathway / C-X-C motif chemokine 12 receptor activity / response to ultrasound / negative regulation of leukocyte tethering or rolling / positive regulation of macrophage migration inhibitory factor signaling pathway / regulation of actin polymerization or depolymerization / chemokine receptor binding / Specification of primordial germ cells / CXCL12-activated CXCR4 signaling pathway ...telencephalon cell migration / chemokine (C-X-C motif) ligand 12 signaling pathway / C-X-C motif chemokine 12 receptor activity / response to ultrasound / negative regulation of leukocyte tethering or rolling / positive regulation of macrophage migration inhibitory factor signaling pathway / regulation of actin polymerization or depolymerization / chemokine receptor binding / Specification of primordial germ cells / CXCL12-activated CXCR4 signaling pathway / myosin light chain binding / myelin maintenance / CXCR chemokine receptor binding / C-X-C chemokine receptor activity / positive regulation of axon extension involved in axon guidance / positive regulation of vasculature development / positive regulation of dopamine secretion / Signaling by ROBO receptors / regulation of chemotaxis / induction of positive chemotaxis / Formation of definitive endoderm / integrin activation / negative regulation of dendritic cell apoptotic process / negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage / C-C chemokine receptor activity / cellular response to chemokine / chemokine-mediated signaling pathway / Developmental Lineage of Pancreatic Acinar Cells / C-C chemokine binding / positive regulation of monocyte chemotaxis / chemokine activity / Chemokine receptors bind chemokines / blood circulation / anchoring junction / dendritic cell chemotaxis / positive regulation of calcium ion import / cellular response to cytokine stimulus / detection of temperature stimulus involved in sensory perception of pain / cell leading edge / positive regulation of oligodendrocyte differentiation / animal organ regeneration / Binding and entry of HIV virion / detection of mechanical stimulus involved in sensory perception of pain / positive regulation of T cell migration / Nuclear signaling by ERBB4 / regulation of cell adhesion / adenylate cyclase inhibitor activity / coreceptor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / positive regulation of endothelial cell proliferation / D2 dopamine receptor binding / response to prostaglandin E / neurogenesis / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / positive regulation of neuron differentiation / adenylate cyclase-inhibiting serotonin receptor signaling pathway / positive regulation of cell adhesion / axon guidance / cellular response to forskolin / regulation of mitotic spindle organization / ubiquitin binding / adult locomotory behavior / cell chemotaxis / growth factor activity / Regulation of insulin secretion / calcium-mediated signaling / defense response / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / brain development / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / integrin binding / response to virus / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / neuron migration / Olfactory Signaling Pathway / Activation of the phototransduction cascade / chemotaxis / intracellular calcium ion homeostasis / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
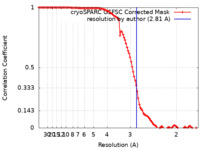
| Method | single particle reconstruction / cryo EM / Resolution: 2.81 Å | |||||||||
 Authors Authors | Liu YZ / Liu AJ / Liao QW / Ye RD | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: Cryo-EM structure of monomeric CXCL12-bound CXCR4 in the active state. Authors: Yezhou Liu / Aijun Liu / Xinyu Li / Qiwen Liao / Weijia Zhang / Lizhe Zhu / Richard D Ye /  Abstract: CXCR4 binding of its endogenous agonist CXCL12 leads to diverse functions, including bone marrow retention of hematopoietic progenitors and cancer metastasis. However, the structure of the CXCL12- ...CXCR4 binding of its endogenous agonist CXCL12 leads to diverse functions, including bone marrow retention of hematopoietic progenitors and cancer metastasis. However, the structure of the CXCL12-bound CXCR4 remains unresolved despite available structures of CXCR4 in complex with antagonists. Here, we present the cryoelectron microscopy (cryo-EM) structure of the CXCL12-CXCR4-Gi complex at an overall resolution of 2.65 Å. CXCL12 forms a 1:1 stoichiometry complex with CXCR4, following the two-site model. The first 8 amino acids of mature CXCL12 are crucial for CXCR4 activation by forming polar interactions with minor sub-pocket residues in the transmembrane binding pocket. The 3.2-Å distance between V3 of CXCL12 and the "toggle switch" W marks the deepest insertion among all chemokine-receptor pairs, leading to conformational changes of CXCR4 for G protein activation. These results, combined with functional assays and computational analysis, provide the structural basis for CXCR4 activation by CXCL12. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36869.map.gz emd_36869.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36869-v30.xml emd-36869-v30.xml emd-36869.xml emd-36869.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36869_fsc.xml emd_36869_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_36869.png emd_36869.png | 50.5 KB | ||
| Filedesc metadata |  emd-36869.cif.gz emd-36869.cif.gz | 7 KB | ||
| Others |  emd_36869_half_map_1.map.gz emd_36869_half_map_1.map.gz emd_36869_half_map_2.map.gz emd_36869_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36869 http://ftp.pdbj.org/pub/emdb/structures/EMD-36869 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36869 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36869 | HTTPS FTP |
-Related structure data
| Related structure data |  8k3zMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36869.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36869.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
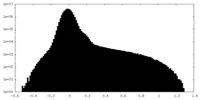
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
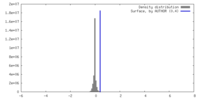
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36869_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
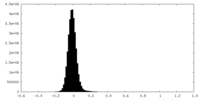
| Density Histograms |
-Half map: #2
| File | emd_36869_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
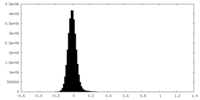
| Density Histograms |
- Sample components
Sample components
-Entire : CXCR4-CXCL12-Gi-scFv16 complex
| Entire | Name: CXCR4-CXCL12-Gi-scFv16 complex |
|---|---|
| Components |
|
-Supramolecule #1: CXCR4-CXCL12-Gi-scFv16 complex
| Supramolecule | Name: CXCR4-CXCL12-Gi-scFv16 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #6, #5, #2-#3, #1, #4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: C-X-C chemokine receptor type 4
| Macromolecule | Name: C-X-C chemokine receptor type 4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.826059 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KEPCFREENA NFNKIFLPTI YSIIFLTGIV GNGLVILVMG YQKKLRSMTD KYRLHLSVAD LLFVITLPFW AVDAVANWYF GNFLCKAVH VIYTVNLYSS VLILAFISLD RYLAIVHATN SQRPRKLLAE KVVYVGVWIP ALLLTIPDFI FANVSEADDR Y ICDRFYPN ...String: KEPCFREENA NFNKIFLPTI YSIIFLTGIV GNGLVILVMG YQKKLRSMTD KYRLHLSVAD LLFVITLPFW AVDAVANWYF GNFLCKAVH VIYTVNLYSS VLILAFISLD RYLAIVHATN SQRPRKLLAE KVVYVGVWIP ALLLTIPDFI FANVSEADDR Y ICDRFYPN DLWVVVFQFQ HIMVGLILPG IVILSCYCII ISKLSHSKGH QKRKALKTTV ILILAFFACW LPYYIGISID SF ILLEIIK QGCEFENTVH KWISITEALA FFHCCLNPIL YAFLGAKFKT SAQHALTSV UniProtKB: C-X-C chemokine receptor type 4 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.198656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLVS ASQDGKLIIW DSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG HTGYLSCCRF LDDNQIVTSS G DTTCALWD ...String: ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLVS ASQDGKLIIW DSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG HTGYLSCCRF LDDNQIVTSS G DTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TFTGHESDIN AICFFPNGNA FA TGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKADRAGVLA GHDNRVSCLG VTD DGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.415031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGGQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #4: Stromal cell-derived factor 1
| Macromolecule | Name: Stromal cell-derived factor 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.292648 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KPVSLSYRCP CRFFESHVAR ANVKHLKILN TPNCALQIVA RLKNNNRQVC IDPKLKWIQE YL UniProtKB: Stromal cell-derived factor 1 |
-Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.131084 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: IAQARKLVEQ LKMEANIDRI KVSKAAADLM AYCEAHAKED PLLTPVPASE NPFRE UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #6: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.337307 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Material: GOLD |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)