-Search query
-Search result
Showing all 48 items for (author: yao & yn)

EMDB-37895: 
PNPase mutant of Mycobacterium tuberculosis

EMDB-37896: 
PNPase of M.tuberculosis with its RNA substrate

EMDB-37905: 
PNPase of Mycobacterium tuberculosis

EMDB-41252: 
Cryo-EM map of the Saccharomyces cerevisiae PCNA clamp unloader Elg1-RFC complex

EMDB-41253: 
Cryo-EM map of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to a cracked PCNA

EMDB-41254: 
Cryo-EM map of the Saccharomyces cerevisiae clamp unloader Elg1-RFC bound to PCNA

PDB-8jgg: 
CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist BAM8-22

EMDB-29578: 
G4 RNA-mediated PRC2 dimer
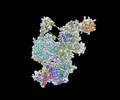
EMDB-29647: 
Body1 of G4 RNA-mediated PRC2 dimer from multibody refinement
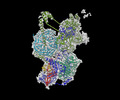
EMDB-29656: 
Body2 of G4 RNA-mediated PRC2 dimer from multibody refinement

PDB-8fyh: 
G4 RNA-mediated PRC2 dimer
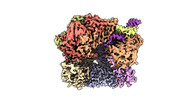
EMDB-29412: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 1 (open 9-1-1 and shoulder bound DNA only)
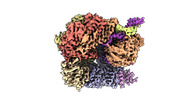
EMDB-29413: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 2 (open 9-1-1 ring and flexibly bound chamber DNA)
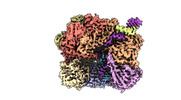
EMDB-29414: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 3 (open 9-1-1 and stably bound chamber DNA)
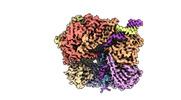
EMDB-29415: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 4 (partially closed 9-1-1 and stably bound chamber DNA)
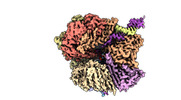
EMDB-29416: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 5 (closed 9-1-1 and stably bound chamber DNA)
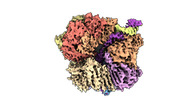
EMDB-29417: 
EM map of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 5-nt gapped DNA (9-1-1 encircling fully bound DNA)

EMDB-27872: 
Cryo-EM structure of the PAC1R-PACAP27-Gs complex

EMDB-27873: 
Cryo-EM structure of the VPAC1R-PACAP27-Gs complex

EMDB-27874: 
Cryo-EM structure of the VPAC1R-VIP-Gs complex

PDB-8e3x: 
Cryo-EM structure of the PAC1R-PACAP27-Gs complex

PDB-8e3y: 
Cryo-EM structure of the VPAC1R-PACAP27-Gs complex

PDB-8e3z: 
Cryo-EM structure of the VPAC1R-VIP-Gs complex

EMDB-25872: 
Cryo-EM 3D map of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to two DNA molecules, one at the 5'-recessed end and the other at the 3'-recessed end

EMDB-25873: 
Cryo-EM 3D map of the S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with an open clamp

EMDB-25874: 
Cryo-EM 3D map of S. cerevisiae clamp-clamp loader complex PCNA-RFC bound to DNA with a closed clamp ring

EMDB-26429: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1520

EMDB-26430: 
Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement

EMDB-26431: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1717

EMDB-26432: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1791

PDB-7uap: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1520

PDB-7uaq: 
Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement

PDB-7uar: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the neutralizing antibody Fab fragment, C1717

EMDB-30406: 
The LAT2-4F2hc complex in complex with tryptophan

EMDB-30407: 
The LAT2-4F2hc complex in complex with leucine

EMDB-21671: 
CryoEM Structure of the glucagon receptor with a dual-agonist peptide

PDB-6whc: 
CryoEM Structure of the glucagon receptor with a dual-agonist peptide

EMDB-0903: 
Heteromeric amino acid transporter b0,+AT-rBAT complex bound with Arginine

EMDB-0904: 
Heteromeric amino acid transporter b0,+AT-rBAT complex

EMDB-0905: 
cryo EM map of b0,+AT-rBAT complex, focused refined on soluble domain

EMDB-0906: 
Cryo EM map of b0,+AT-rBAT complex, focused refined on TM domain

EMDB-0907: 
Cryo EM map of b0,+AT-rBAT complex bound with Arginine, focused refined on soluble domain

EMDB-0908: 
Cryo EM map of b0,+ AT-rBAT complex bound with Arginine, focused refined on TM domain

EMDB-20074: 
Chimpanzee SIV Env trimeric ectodomain.

PDB-6ohy: 
Chimpanzee SIV Env trimeric ectodomain.

PDB-3j9b: 
Electron cryo-microscopy of an RNA polymerase

EMDB-6202: 
Electron cryo-microscopy of an RNA polymerase

EMDB-6203: 
Electron cryo-microscopy of an RNA polymerase
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
