+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0904 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Heteromeric amino acid transporter b0,+AT-rBAT complex | ||||||||||||||||||||||||
 Map data Map data | Cryo EM map of b0, AT-rBAT complex | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | transporter / MEMBRANE PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationbroad specificity neutral L-amino acid:basic L-amino acid antiporter activity / Defective SLC3A1 causes cystinuria (CSNU) / Defective SLC7A9 causes cystinuria (CSNU) / basic amino acid transport / basic amino acid transmembrane transporter activity / L-cystine transmembrane transporter activity / L-cystine transport / amino acid transmembrane transport / aspartate transmembrane transport / L-glutamate transmembrane transport ...broad specificity neutral L-amino acid:basic L-amino acid antiporter activity / Defective SLC3A1 causes cystinuria (CSNU) / Defective SLC7A9 causes cystinuria (CSNU) / basic amino acid transport / basic amino acid transmembrane transporter activity / L-cystine transmembrane transporter activity / L-cystine transport / amino acid transmembrane transport / aspartate transmembrane transport / L-glutamate transmembrane transport / neutral amino acid transport / neutral L-amino acid transmembrane transporter activity / Amino acid transport across the plasma membrane / amino acid transmembrane transporter activity / vacuolar membrane / antiporter activity / Basigin interactions / amino acid transport / brush border membrane / peptide antigen binding / protein-containing complex assembly / gene expression / carbohydrate metabolic process / apical plasma membrane / protein heterodimerization activity / protein-containing complex binding / extracellular exosome / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | ||||||||||||||||||||||||
 Authors Authors | Yan RH / Li YN | ||||||||||||||||||||||||
| Funding support |  China, 7 items China, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2020 Journal: Sci Adv / Year: 2020Title: Cryo-EM structure of the human heteromeric amino acid transporter bAT-rBAT. Authors: Renhong Yan / Yaning Li / Yi Shi / Jiayao Zhou / Jianlin Lei / Jing Huang / Qiang Zhou /  Abstract: Heteromeric amino acid transporters (HATs) catalyze the transmembrane movement of amino acids, comprising two subunits, a heavy chain and a light chain, linked by a disulfide bridge. The bAT (SLC7A9) ...Heteromeric amino acid transporters (HATs) catalyze the transmembrane movement of amino acids, comprising two subunits, a heavy chain and a light chain, linked by a disulfide bridge. The bAT (SLC7A9) is a representative light chain of HATs, forming heterodimer with rBAT, a heavy chain which mediates the membrane trafficking of bAT. The bAT-rBAT complex is an obligatory exchanger, which mediates the influx of cystine and cationic amino acids and the efflux of neutral amino acids in kidney and small intestine. Here, we report the cryo-EM structure of the human bAT-rBAT complex alone and in complex with arginine substrate at resolution of 2.7 and 2.3 Å, respectively. The overall structure of bAT-rBAT exists as a dimer of heterodimer consistent with the previous study. A ligand molecule is bound to the substrate binding pocket, near which an occluded pocket is identified, to which we found that it is important for substrate transport. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0904.map.gz emd_0904.map.gz | 116.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0904-v30.xml emd-0904-v30.xml emd-0904.xml emd-0904.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0904.png emd_0904.png | 44.5 KB | ||
| Filedesc metadata |  emd-0904.cif.gz emd-0904.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0904 http://ftp.pdbj.org/pub/emdb/structures/EMD-0904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0904 | HTTPS FTP |
-Related structure data
| Related structure data |  6lidMC  0903C  0905C  0906C  0907C  0908C  6li9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0904.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0904.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo EM map of b0, AT-rBAT complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.091 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
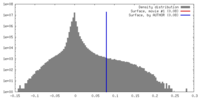
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : b0,+AT-rBAT complex
| Entire | Name: b0,+AT-rBAT complex |
|---|---|
| Components |
|
-Supramolecule #1: b0,+AT-rBAT complex
| Supramolecule | Name: b0,+AT-rBAT complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: Neutral and basic amino acid transport protein rBAT
| Macromolecule | Name: Neutral and basic amino acid transport protein rBAT / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 80.691586 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAHHHHHHHH HHSGRAEDKS KRDSIEMSMK GCQTNNGFVH NEDILEQTPD PGSSTDNLKH STRGILGSQE PDFKGVQPYA GMPKEVLFQ FSGQARYRIP REILFWLTVA SVLVLIAATI AIIALSPKCL DWWQEGPMYQ IYPRSFKDSN KDGNGDLKGI Q DKLDYITA ...String: MAHHHHHHHH HHSGRAEDKS KRDSIEMSMK GCQTNNGFVH NEDILEQTPD PGSSTDNLKH STRGILGSQE PDFKGVQPYA GMPKEVLFQ FSGQARYRIP REILFWLTVA SVLVLIAATI AIIALSPKCL DWWQEGPMYQ IYPRSFKDSN KDGNGDLKGI Q DKLDYITA LNIKTVWITS FYKSSLKDFR YGVEDFREVD PIFGTMEDFE NLVAAIHDKG LKLIIDFIPN HTSDKHIWFQ LS RTRTGKY TDYYIWHDCT HENGKTIPPN NWLSVYGNSS WHFDEVRNQC YFHQFMKEQP DLNFRNPDVQ EEIKEILRFW LTK GVDGFS LDAVKFLLEA KHLRDEIQVN KTQIPDTVTQ YSELYHDFTT TQVGMHDIVR SFRQTMDQYS TEPGRYRFMG TEAY AESID RTVMYYGLPF IQEADFPFNN YLSMLDTVSG NSVYEVITSW MENMPEGKWP NWMIGGPDSS RLTSRLGNQY VNVMN MLLF TLPGTPITYY GEEIGMGNIV AANLNESYDI NTLRSKSPMQ WDNSSNAGFS EASNTWLPTN SDYHTVNVDV QKTQPR SAL KLYQDLSLLH ANELLLNRGW FCHLRNDSHY VVYTRELDGI DRIFIVVLNF GESTLLNLHN MISGLPAKMR IRLSTNS AD KGSKVDTSGI FLDKGEGLIF EHNTKNLLHR QTAFRDRCFV SNRACYSSVL NILYTSC UniProtKB: Amino acid transporter heavy chain SLC3A1 |
-Macromolecule #2: b(0,+)-type amino acid transporter 1
| Macromolecule | Name: b(0,+)-type amino acid transporter 1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.656105 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADYKDDDDK SGPDEVDASG RGDTGLRKRR EDEKSIQSQE PKTTSLQKEL GLISGISIIV GTIIGSGIFV SPKSVLSNTE AVGPCLIIW AACGVLATLG ALCFAELGTM ITKSGGEYPY LMEAYGPIPA YLFSWASLIV IKPTSFAIIC LSFSEYVCAP F YVGCKPPQ ...String: MADYKDDDDK SGPDEVDASG RGDTGLRKRR EDEKSIQSQE PKTTSLQKEL GLISGISIIV GTIIGSGIFV SPKSVLSNTE AVGPCLIIW AACGVLATLG ALCFAELGTM ITKSGGEYPY LMEAYGPIPA YLFSWASLIV IKPTSFAIIC LSFSEYVCAP F YVGCKPPQ IVVKCLAAAA ILFISTVNSL SVRLGSYVQN IFTAAKLVIV AIIIISGLVL LAQGNTKNFD NSFEGAQLSV GA ISLAFYN GLWAYDGWNQ LNYITEELRN PYRNLPLAII IGIPLVTACY ILMNVSYFTV MTATELLQSQ AVAVTFGDRV LYP ASWIVP LFVAFSTIGA ANGTCFTAGR LIYVAGREGH MLKVLSYISV RRLTPAPAII FYGIIATIYI IPGDINSLVN YFSF AAWLF YGLTILGLIV MRFTRKELER PIKVPVVIPV LMTLISVFLV LAPIISKPTW EYLYCVLFIL SGLLFYFLFV HYKFG WAQK ISKPITMHLQ MLMEVVPPEE DPE UniProtKB: b(0,+)-type amino acid transporter 1 |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #5: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE
| Macromolecule | Name: 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE / type: ligand / ID: 5 / Number of copies: 4 / Formula: 3PH |
|---|---|
| Molecular weight | Theoretical: 704.998 Da |
| Chemical component information |  ChemComp-3PH: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 246 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3) / Number images used: 127377 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)