-Search query
-Search result
Showing 1 - 50 of 3,776 items for (author: xu & q)

EMDB entry, No image
EMDB-36782: 
SID1 transmembrane family member 2

EMDB entry, No image
EMDB-36783: 
SID1 transmembrane family member 2

EMDB entry, No image
EMDB-36784: 
SID1 transmembrane family member 2

EMDB entry, No image
EMDB-36785: 
SID1 transmembrane family member 1

EMDB entry, No image
EMDB-36791: 
SID1 transmembrane family member 1

EMDB entry, No image
EMDB-36792: 
SID1 transmembrane family member 1

PDB-8k10: 
SID1 transmembrane family member 2

PDB-8k11: 
SID1 transmembrane family member 2

PDB-8k12: 
SID1 transmembrane family member 2

PDB-8k13: 
SID1 transmembrane family member 1

PDB-8k1b: 
SID1 transmembrane family member 1

PDB-8k1d: 
SID1 transmembrane family member 1

EMDB-37105: 
ATP-bound hMRP5 outward-open

EMDB-37554: 
wt-hMRP5 inward-open

EMDB-37555: 
RD-hMRP5-inward open

EMDB-37556: 
ND-hMRP5-inward open

EMDB-37557: 
m6-hMRP5 inward open

EMDB-37558: 
M5PI-bound hMRP5

PDB-8kci: 
ATP-bound hMRP5 outward-open

PDB-8wi0: 
wt-hMRP5 inward-open

PDB-8wi2: 
RD-hMRP5-inward open

PDB-8wi3: 
ND-hMRP5-inward open

PDB-8wi4: 
m6-hMRP5 inward open

PDB-8wi5: 
M5PI-bound hMRP5

EMDB-38502: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P) in complex with human ACE2

EMDB-38460: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P), 1-RBD-up state

EMDB-38463: 
Cryo-EM structure of SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state

EMDB-38476: 
Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P), RBD-closed state

EMDB-38488: 
Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P), RBD-closed state

EMDB-38495: 
SARS-CoV-2 Omicron EG.5.1 RBD in complex with human ACE2 (local refined from the spike protein)

EMDB-38496: 
SARS-CoV-2 Omicron HV.1 RBD in complex with human ACE2 (local refinement from the spike protein)

EMDB-38498: 
Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P) in complex with human ACE2

EMDB-38505: 
Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P) in complex with human ACE2

EMDB-37711: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2
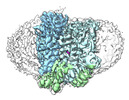
EMDB-38966: 
Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate
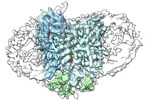
EMDB-38968: 
Cryo-EM structure of human urate transporter GLUT9 bound to inhibitor apigenin

PDB-8y65: 
Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate

PDB-8y66: 
Cryo-EM structure of human urate transporter GLUT9 bound to inhibitor apigenin

EMDB-37210: 
Prefusion RSV F Bound to Lonafarnib and D25 Fab

PDB-8kg5: 
Prefusion RSV F Bound to Lonafarnib and D25 Fab

EMDB-37944: 
Structure of 26RFa-pyroglutamylated RFamide peptide receptor complex

PDB-8wz2: 
Structure of 26RFa-pyroglutamylated RFamide peptide receptor complex

EMDB-41569: 
Cryo-EM structure of HmAb64 scFv in complex with CNE40 SOSIP trimer

PDB-8tr3: 
Cryo-EM structure of HmAb64 scFv in complex with CNE40 SOSIP trimer

EMDB-37823: 
Cryo-EM structure of Melanin-Concentrating Hormone Receptor 1 with MCH

EMDB-37824: 
Cryo-EM structure of Melanin-Concentrating Hormone Receptor 2 with MCH

PDB-8wss: 
Cryo-EM structure of Melanin-Concentrating Hormone Receptor 1 with MCH

PDB-8wst: 
Cryo-EM structure of Melanin-Concentrating Hormone Receptor 2 with MCH

EMDB-37756: 
Cryo-EM structure of bsAb3 Fab-Gn-Gc complex

PDB-8wqw: 
Cryo-EM structure of bsAb3 Fab-Gn-Gc complex
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
