[English] 日本語
 Yorodumi
Yorodumi- EMDB-38463: Cryo-EM structure of SARS-CoV-2 Omicron EG.5 spike protein(6P), R... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / Omicron / EG.5 / spike protein / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.61 Å | |||||||||
 Authors Authors | Li LJ / Gu YH / Shi KY / Qi JX / Gao GF | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants. Authors: Linjie Li / Kaiyuan Shi / Yuhang Gu / Zepeng Xu / Chang Shu / Dedong Li / Junqing Sun / Mengqing Cong / Xiaomei Li / Xin Zhao / Guanghui Yu / Songnian Hu / Hui Tan / Jianxun Qi / Xiaopeng Ma ...Authors: Linjie Li / Kaiyuan Shi / Yuhang Gu / Zepeng Xu / Chang Shu / Dedong Li / Junqing Sun / Mengqing Cong / Xiaomei Li / Xin Zhao / Guanghui Yu / Songnian Hu / Hui Tan / Jianxun Qi / Xiaopeng Ma / Kefang Liu / George F Gao /  Abstract: The recently emerged BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 variants have a growth advantage. In this study, we explore the structural bases of receptor binding and immune evasion for the Omicron BA.2. ...The recently emerged BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 variants have a growth advantage. In this study, we explore the structural bases of receptor binding and immune evasion for the Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants. Our findings reveal that BA.2.86 exhibits strong receptor binding, whereas its JN.1 sub-lineage displays a decreased binding affinity to human ACE2 (hACE2). Through complex structure analyses, we observed that the reversion of R493Q in BA.2.86 receptor binding domain (RBD) plays a facilitating role in receptor binding, while the L455S substitution in JN.1 RBD restores optimal affinity. Furthermore, the structure of monoclonal antibody (mAb) S309 complexed with BA.2.86 RBD highlights the importance of the K356T mutation, which brings a new N-glycosylation motif, altering the binding pattern of mAbs belonging to RBD-5 represented by S309. These findings emphasize the importance of closely monitoring BA.2.86 and its sub-lineages to prevent another wave of SARS-CoV-2 infections. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38463.map.gz emd_38463.map.gz | 449.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38463-v30.xml emd-38463-v30.xml emd-38463.xml emd-38463.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38463.png emd_38463.png | 96.1 KB | ||
| Masks |  emd_38463_msk_1.map emd_38463_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-38463.cif.gz emd-38463.cif.gz | 6.3 KB | ||
| Others |  emd_38463_half_map_1.map.gz emd_38463_half_map_1.map.gz emd_38463_half_map_2.map.gz emd_38463_half_map_2.map.gz | 475.6 MB 475.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38463 http://ftp.pdbj.org/pub/emdb/structures/EMD-38463 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38463 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38463 | HTTPS FTP |
-Related structure data
| Related structure data |  8xm5MC  8wp8C  8xlvC  8xmgC  8xmtC  8xn2C  8xn3C  8xn5C  8xnfC  8xnkC  8y16C  8y18C  8y5jC  8y6aC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38463.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38463.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.808 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_38463_msk_1.map emd_38463_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
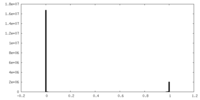
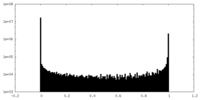
| Density Histograms |
-Half map: #2
| File | emd_38463_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
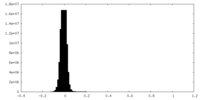
| Density Histograms |
-Half map: #1
| File | emd_38463_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
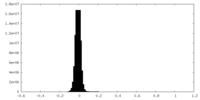
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state
| Entire | Name: SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state
| Supramolecule | Name: SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Details: Omicron EG.5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 141.06675 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTQCVNLITR TQSYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGT NGTKRFDNPA LPFNDGVYFA STEKSNIIRG WIFGTTLDSK TQSLLIVNNA TNVVIKVCEF QFCNDPFLDV Y QKNNKSWM ...String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTQCVNLITR TQSYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGT NGTKRFDNPA LPFNDGVYFA STEKSNIIRG WIFGTTLDSK TQSLLIVNNA TNVVIKVCEF QFCNDPFLDV Y QKNNKSWM ESEFRVYSSA NNCTFEYVSQ PFLMDLEGKE GNFKNLREFV FKNIDGYFKI YSKHTPINLE RDLPQGFSAL EP LVDLPIG INITRFQTLL ALHRSYLTPV DSSSGWTAGA AAYYVGYLQP RTFLLKYNEN GTITDAVDCA LDPLSETKCT LKS FTVEKG IYQTSNFRVQ PTESIVRFPN ITNLCPFHEV FNATTFASVY AWNRKRISNC VADYSVIYNF APFFAFKCYG VSPT KLNDL CFTNVYADSF VIRGNEVSQI APGQTGNIAD YNYKLPDDFT GCVIAWNSNK LDSKPSGNYN YLYRLLRKSK LKPFE RDIS TEIYQAGNKP CNGVAGPNCY SPLQSYGFRP TYGVGHQPYR VVVLSFELLH APATVCGPKK STNLVKNKCV NFNFNG LTG TGVLTESNKK FLPFQQFGRD IADTTDAVRD PQTLEILDIT PCSFGGVSVI TPGTNTSNQV AVLYQGVNCT EVPVAIH AD QLTPTWRVYS TGSNVFQTRA GCLIGAEYVN NSYECDIPIG AGICASYQTQ TKSHRRARSV ASQSIIAYTM SLGAENSV A YSNNSIAIPT NFTISVTTEI LPVSMTKTSV DCTMYICGDS TECSNLLLQY GSFCTQLKRA LTGIAVEQDK NTQEVFAQV KQIYKTPPIK YFGGFNFSQI LPDPSKPSKR SPIEDLLFNK VTLADAGFIK QYGDCLGDIA ARDLICAQKF NGLTVLPPLL TDEMIAQYT SALLAGTITS GWTFGAGPAL QIPFPMQMAY RFNGIGVTQN VLYENQKLIA NQFNSAIGKI QDSLSSTPSA L GKLQDVVN HNAQALNTLV KQLSSKFGAI SSVLNDILSR LDPPEAEVQI DRLITGRLQS LQTYVTQQLI RAAEIRASAN LA ATKMSEC VLGQSKRVDF CGKGYHLMSF PQSAPHGVVF LHVTYVPAQE KNFTTAPAIC HDGKAHFPRE GVFVSNGTHW FVT QRNFYE PQIITTDNTF VSGNCDVVIG IVNNTVYDPL QPELDSFKEE LDKYFKNHTS PDVDLGDISG INASVVNIQK EIDR LNEVA KNLNESLIDL QELGKYEQGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGSA WSHPQFEKHH HHHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 27 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.61 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 473611 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)