[English] 日本語
 Yorodumi
Yorodumi- EMDB-38966: Cryo-EM structure of human urate transporter GLUT9 bound to subst... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate | |||||||||
 Map data Map data | Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate in detergent micelles | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GLUT9 / Urate / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC2A9 causes hypouricemia renal 2 (RHUC2) / fructose transmembrane transporter activity / fructose transmembrane transport / dehydroascorbic acid transport / hexose transmembrane transport / carbohydrate:proton symporter activity / Cellular hexose transport / D-glucose transmembrane transporter activity / D-glucose transmembrane transport / urate transport ...Defective SLC2A9 causes hypouricemia renal 2 (RHUC2) / fructose transmembrane transporter activity / fructose transmembrane transport / dehydroascorbic acid transport / hexose transmembrane transport / carbohydrate:proton symporter activity / Cellular hexose transport / D-glucose transmembrane transporter activity / D-glucose transmembrane transport / urate transport / urate metabolic process / urate transmembrane transporter activity / : / transmembrane transporter activity / basolateral plasma membrane / apical plasma membrane / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
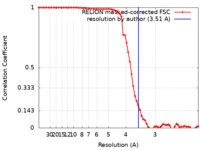
| Method | single particle reconstruction / cryo EM / Resolution: 3.51 Å | |||||||||
 Authors Authors | Pan XJ / Shen ZL / Xu L / Huang GXY | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for urate recognition and apigenin inhibition of human GLUT9. Authors: Zilin Shen / Li Xu / Tong Wu / Huan Wang / Qifan Wang / Xiaofei Ge / Fang Kong / Gaoxingyu Huang / Xiaojing Pan /  Abstract: Urate, the physiological form of uric acid and a potent antioxidant in serum, plays a pivotal role in scavenging reactive oxygen species. Yet excessive accumulation of urate, known as hyperuricemia, ...Urate, the physiological form of uric acid and a potent antioxidant in serum, plays a pivotal role in scavenging reactive oxygen species. Yet excessive accumulation of urate, known as hyperuricemia, is the primary risk factor for the development of gout. The high-capacity urate transporter GLUT9 represents a promising target for gout treatment. Here, we present cryo-electron microscopy structures of human GLUT9 in complex with urate or its inhibitor apigenin at overall resolutions of 3.5 Å and 3.3 Å, respectively. In both structures, GLUT9 exhibits an inward open conformation, wherein the substrate binding pocket faces the intracellular side. These structures unveil the molecular basis for GLUT9's substrate preference of urate over glucose, and show that apigenin acts as a competitive inhibitor by occupying the substrate binding site. Our findings provide critical information for the development of specific inhibitors targeting GLUT9 as potential therapeutics for gout and hyperuricemia. | |||||||||
| History |
|
- Structure visualization
Structure visualization
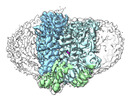
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38966.map.gz emd_38966.map.gz | 25.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38966-v30.xml emd-38966-v30.xml emd-38966.xml emd-38966.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38966_fsc.xml emd_38966_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_38966.png emd_38966.png | 155.8 KB | ||
| Filedesc metadata |  emd-38966.cif.gz emd-38966.cif.gz | 5.5 KB | ||
| Others |  emd_38966_half_map_1.map.gz emd_38966_half_map_1.map.gz emd_38966_half_map_2.map.gz emd_38966_half_map_2.map.gz | 25 MB 25 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38966 http://ftp.pdbj.org/pub/emdb/structures/EMD-38966 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38966 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38966 | HTTPS FTP |
-Related structure data
| Related structure data |  8y65MC  8y66C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38966.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38966.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate in detergent micelles | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0979 Å | ||||||||||||||||||||||||||||||||||||
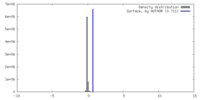
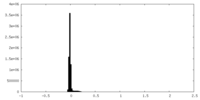
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38966_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
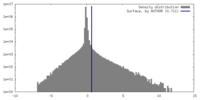
| Density Histograms |
-Half map: #1
| File | emd_38966_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
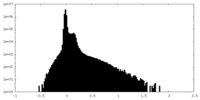
| Density Histograms |
- Sample components
Sample components
-Entire : GLUT9 in complex with urate
| Entire | Name: GLUT9 in complex with urate |
|---|---|
| Components |
|
-Supramolecule #1: GLUT9 in complex with urate
| Supramolecule | Name: GLUT9 in complex with urate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 2, facilitated glucose transporter member 9
| Macromolecule | Name: Solute carrier family 2, facilitated glucose transporter member 9 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 63.21509 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASWSHPQFE KGGGARGGSG GGSWSHPQFE KGFDYKDDDD KGTARKQNRN SKELGLVPLT DDTSHAGPPG PGRALLECDH LRSGVPGGR RRKDWSCSLL VASLAGAFGS SFLYGYNLSV VNAPTPYIKA FYNESWERRH GRPIDPDTLT LLWSVTVSIF A IGGLVGTL ...String: MASWSHPQFE KGGGARGGSG GGSWSHPQFE KGFDYKDDDD KGTARKQNRN SKELGLVPLT DDTSHAGPPG PGRALLECDH LRSGVPGGR RRKDWSCSLL VASLAGAFGS SFLYGYNLSV VNAPTPYIKA FYNESWERRH GRPIDPDTLT LLWSVTVSIF A IGGLVGTL IVKMIGKVLG RKHTLLANNG FAISAALLMA CSLQAGAFEM LIVGRFIMGI DGGVALSVLP MYLSEISPKE IR GSLGQVT AIFICIGVFT GQLLGLPELL GKESTWPYLF GVIVVPAVVQ LLSLPFLPDS PRYLLLEKHN EARAVKAFQT FLG KADVSQ EVEEVLAESR VQRSIRLVSV LELLRAPYVR WQVVTVIVTM ACYQLCGLNA IWFYTNSIFG KAGIPPAKIP YVTL STGGI ETLAAVFSGL VIEHLGRRPL LIGGFGLMGL FFGTLTITLT LQDHAPWVPY LSIVGILAII ASFCSGPGGI PFILT GEFF QQSQRPAAFI IAGTVNWLSN FAVGLLFPFI QKSLDTYCFL VFATICITGA IYLYFVLPET KNRTYAEISQ AFSKRN KAY PPEEKIDSAV TDGKINGRP UniProtKB: Solute carrier family 2, facilitated glucose transporter member 9 |
-Macromolecule #2: URIC ACID
| Macromolecule | Name: URIC ACID / type: ligand / ID: 2 / Number of copies: 1 / Formula: URC |
|---|---|
| Molecular weight | Theoretical: 168.11 Da |
| Chemical component information |  ChemComp-URC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 6 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 2.56 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)