+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | wt-hMRP5 inward-open | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | wt-hMRP5 inward-open / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpurine nucleotide transport / macromolecule transmembrane transporter activity / cGMP transport / carbohydrate derivative transmembrane transporter activity / : / folate transmembrane transport / purine nucleotide transmembrane transporter activity / cAMP transport / heme transmembrane transporter activity / hyaluronan biosynthetic process ...purine nucleotide transport / macromolecule transmembrane transporter activity / cGMP transport / carbohydrate derivative transmembrane transporter activity / : / folate transmembrane transport / purine nucleotide transmembrane transporter activity / cAMP transport / heme transmembrane transporter activity / hyaluronan biosynthetic process / heme transmembrane transport / glutathione transmembrane transport / glutathione transmembrane transporter activity / : / xenobiotic transmembrane transport / : / xenobiotic detoxification by transmembrane export across the plasma membrane / Paracetamol ADME / export across plasma membrane / ABC-type xenobiotic transporter / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / Azathioprine ADME / ABC-type xenobiotic transporter activity / xenobiotic transport / efflux transmembrane transporter activity / ATPase-coupled transmembrane transporter activity / xenobiotic transmembrane transporter activity / transport across blood-brain barrier / xenobiotic metabolic process / ABC-family proteins mediated transport / transmembrane transport / Golgi lumen / basolateral plasma membrane / endosome membrane / apical plasma membrane / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.93 Å | |||||||||
 Authors Authors | Liu ZM / Huang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Inhibition and transport mechanisms of the ABC transporter hMRP5. Authors: Ying Huang / Chenyang Xue / Ruiqian Bu / Cang Wu / Jiachen Li / Jinqiu Zhang / Jinyu Chen / Zhaoying Shi / Yonglong Chen / Yong Wang / Zhongmin Liu /  Abstract: Human multidrug resistance protein 5 (hMRP5) effluxes anticancer and antivirus drugs, driving multidrug resistance. To uncover the mechanism of hMRP5, we determine six distinct cryo-EM structures, ...Human multidrug resistance protein 5 (hMRP5) effluxes anticancer and antivirus drugs, driving multidrug resistance. To uncover the mechanism of hMRP5, we determine six distinct cryo-EM structures, revealing an autoinhibitory N-terminal peptide that must dissociate to permit subsequent substrate recruitment. Guided by these molecular insights, we design an inhibitory peptide that could block substrate entry into the transport pathway. We also identify a regulatory motif, comprising a positively charged cluster and hydrophobic patches, within the first nucleotide-binding domain that modulates hMRP5 localization by engaging with membranes. By integrating our structural, biochemical, computational, and cell biological findings, we propose a model for hMRP5 conformational cycling and localization. Overall, this work provides mechanistic understanding of hMRP5 function, while informing future selective hMRP5 inhibitor development. More broadly, this study advances our understanding of the structural dynamics and inhibition of ABC transporters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37554.map.gz emd_37554.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37554-v30.xml emd-37554-v30.xml emd-37554.xml emd-37554.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37554.png emd_37554.png | 58.6 KB | ||
| Filedesc metadata |  emd-37554.cif.gz emd-37554.cif.gz | 6.7 KB | ||
| Others |  emd_37554_half_map_1.map.gz emd_37554_half_map_1.map.gz emd_37554_half_map_2.map.gz emd_37554_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37554 http://ftp.pdbj.org/pub/emdb/structures/EMD-37554 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37554 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37554 | HTTPS FTP |
-Related structure data
| Related structure data |  8wi0MC  8kciC  8wi2C  8wi3C  8wi4C  8wi5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37554.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37554.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.072 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37554_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
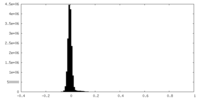
| Density Histograms |
-Half map: #1
| File | emd_37554_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
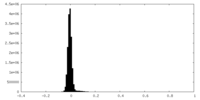
| Density Histograms |
- Sample components
Sample components
-Entire : wt-hMRP5
| Entire | Name: wt-hMRP5 |
|---|---|
| Components |
|
-Supramolecule #1: wt-hMRP5
| Supramolecule | Name: wt-hMRP5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ATP-binding cassette sub-family C member 5
| Macromolecule | Name: ATP-binding cassette sub-family C member 5 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 160.86375 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKDIDIGKEY IIPSPGYRSV RERTSTSGTH RDREDSKFRR TRPLECQDAL ETAARAEGLS LDASMHSQLR ILDEEHPKGK YHHGLSALK PIRTTSKHQH PVDNAGLFSC MTFSWLSSLA RVAHKKGELS MEDVWSLSKH ESSDVNCRRL ERLWQEELNE V GPDAASLR ...String: MKDIDIGKEY IIPSPGYRSV RERTSTSGTH RDREDSKFRR TRPLECQDAL ETAARAEGLS LDASMHSQLR ILDEEHPKGK YHHGLSALK PIRTTSKHQH PVDNAGLFSC MTFSWLSSLA RVAHKKGELS MEDVWSLSKH ESSDVNCRRL ERLWQEELNE V GPDAASLR RVVWIFCRTR LILSIVCLMI TQLAGFSGPA FMVKHLLEYT QATESNLQYS LLLVLGLLLT EIVRSWSLAL TW ALNYRTG VRLRGAILTM AFKKILKLKN IKEKSLGELI NICSNDGQRM FEAAAVGSLL AGGPVVAILG MIYNVIILGP TGF LGSAVF ILFYPAMMFA SRLTAYFRRK CVAATDERVQ KMNEVLTYIK FIKMYAWVKA FSQSVQKIRE EERRILEKAG YFQS ITVGV APIVVVIASV VTFSVHMTLG FDLTAAQAFT VVTVFNSMTF ALKVTPFSVK SLSEASVAVD RFKSLFLMEE VHMIK NKPA SPHIKIEMKN ATLAWDSSHS SIQNSPKLTP KMKKDKRASR GKKEKVRQLQ RTEHQAVLAE QKGHLLLDSD ERPSPE EEE GKHIHLGHLR LQRTLHSIDL EIQEGKLVGI CGSVGSGKTS LISAILGQMT LLEGSIAISG TFAYVAQQAW ILNATLR DN ILFGKEYDEE RYNSVLNSCC LRPDLAILPS SDLTEIGERG ANLSGGQRQR ISLARALYSD RSIYILDDPL SALDAHVG N HIFNSAIRKH LKSKTVLFVT HQLQYLVDCD EVIFMKEGCI TERGTHEELM NLNGDYATIF NNLLLGETPP VEINSKKET SGSQKKSQDK GPKTGSVKKE KAVKPEEGQL VQLEEKGQGS VPWSVYGVYI QAAGGPLAFL VIMALFMLNV GSTAFSTWWL SYWIKQGSG NTTVTRGNET SVSDSMKDNP HMQYYASIYA LSMAVMLILK AIRGVVFVKG TLRASSRLHD ELFRRILRSP M KFFDTTPT GRILNRFSKD MDEVDVRLPF QAEMFIQNVI LVFFCVGMIA GVFPWFLVAV GPLVILFSVL HIVSRVLIRE LK RLDNITQ SPFLSHITSS IQGLATIHAY NKGQEFLHRY QELLDDNQAP FFLFTCAMRW LAVRLDLISI ALITTTGLMI VLM HGQIPP AYAGLAISYA VQLTGLFQFT VRLASETEAR FTSVERINHY IKTLSLEAPA RIKNKAPSPD WPQEGEVTFE NAEM RYREN LPLVLKKVSF TIKPKEKIGI VGRTGSGKSS LGMALFRLVE LSGGCIKIDG VRISDIGLAD LRSKLSIIPQ EPVLF SGTV RSNLDPFNQY TEDQIWDALE RTHMKECIAQ LPLKLESEVM ENGDNFSVGE RQLLCIARAL LRHCKILILD EATAAM DTE TDLLIQETIR EAFADCTMLT IAHRLHTVLG SDRIMVLAQG QVVEFDTPSV LLSNDSSRFY AMFAAAENKV AVKG UniProtKB: ATP-binding cassette sub-family C member 5 |
-Macromolecule #2: ATP-binding cassette sub-family C member 5
| Macromolecule | Name: ATP-binding cassette sub-family C member 5 / type: protein_or_peptide / ID: 2 / Details: peptide:CQDALETAARAEGL(SEP)LDAS / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.002036 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: CQDALETAAR AEGL(SEP)LDAS UniProtKB: ATP-binding cassette sub-family C member 5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)