-Search query
-Search result
Showing 1 - 50 of 156 items for (author: wilk & p)

EMDB-43762: 
Aca2 from Pectobacterium phage ZF40 bound to RNA

PDB-8w35: 
Aca2 from Pectobacterium phage ZF40 bound to RNA
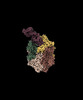
EMDB-18784: 
CryoEM structure of DHS-eIF5A complex structure from Trichomonas vaginalis

PDB-8qzx: 
CryoEM structure of DHS-eIF5A complex structure from Trichomonas vaginalis

EMDB-42880: 
State 1 Yeast V-ATPase with Oxr1p bound

EMDB-42881: 
State 2 Yeast V-ATPase without Oxr1p bound

EMDB-42812: 
PNMA2 capsid, overall icosahedral map

EMDB-42815: 
PNMA2 capsid, focussed refinement of a pentamer (C5 symmetry)

PDB-8uyo: 
Structure of a recombinant human PNMA2 capsid

EMDB-15696: 
IAPP S20G lag-phase fibril polymorph 2PF-P

EMDB-15728: 
IAPP S20G growth-phase fibril polymorph 2PF-L

EMDB-15729: 
IAPP S20G growth-phase fibril polymorph 2PF-C

EMDB-15730: 
IAPP S20G growth-phase fibril polymorph 3PF-CU

EMDB-15731: 
IAPP S20G growth-phase fibril polymorph 4PF-CU

EMDB-15753: 
IAPP S20G plateau-phase fibril polymorph 2PF-L

EMDB-15754: 
IAPP S20G plateau-phase fibril polymorph 4PF-CU

EMDB-15755: 
IAPP S20G plateau-phase fibril polymorph 4PF-LU

EMDB-15756: 
IAPP S20G plateau-phase fibril polymorph 4PF-LJ

PDB-8awt: 
IAPP S20G lag-phase fibril polymorph 2PF-P

PDB-8az0: 
IAPP S20G growth-phase fibril polymorph 2PF-L

PDB-8az1: 
IAPP S20G growth-phase fibril polymorph 2PF-C

PDB-8az2: 
IAPP S20G growth-phase fibril polymorph 3PF-CU
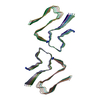
PDB-8az3: 
IAPP S20G growth-phase fibril polymorph 4PF-CU

PDB-8az4: 
IAPP S20G plateau-phase fibril polymorph 2PF-L

PDB-8az5: 
IAPP S20G plateau-phase fibril polymorph 4PF-CU

PDB-8az6: 
IAPP S20G plateau-phase fibril polymorph 4PF-LU

PDB-8az7: 
IAPP S20G plateau-phase fibril polymorph 4PF-LJ

EMDB-16964: 
MutSbeta bound to (CAG)2 DNA (canonical form)

PDB-8olx: 
MutSbeta bound to (CAG)2 DNA (canonical form)

EMDB-16972: 
MutSbeta bound to 61bp homoduplex DNA

PDB-8oma: 
MutSbeta bound to 61bp homoduplex DNA

EMDB-16971: 
MutSbeta bound to (CAG)2 DNA (open form)

PDB-8om9: 
MutSbeta bound to (CAG)2 DNA (open form)

EMDB-16973: 
DNA-unbound MutSbeta-ATP complex (bent clamp form)

PDB-8omo: 
DNA-unbound MutSbeta-ATP complex (bent clamp form)

EMDB-16974: 
DNA-unbound MutSbeta-ATP complex (straight clamp form)

PDB-8omq: 
DNA-unbound MutSbeta-ATP complex (straight clamp form)

EMDB-16975: 
MutSbeta bound to homoduplex plasmid DNA

EMDB-16969: 
DNA-free open form of MutSbeta

PDB-8om5: 
DNA-free open form of MutSbeta

EMDB-15052: 
CryoEM structure of DHS-eIF5A1 complex

PDB-8a0e: 
CryoEM structure of DHS-eIF5A1 complex

EMDB-26809: 
KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate

EMDB-26810: 
KDM2B-nucleosome complex stabilized by H3K36C-UNC8015 covalent conjugate

PDB-7uv9: 
KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate

EMDB-26723: 
Structure of IsrB ternary complex with RNA mutant and target DNA

EMDB-27533: 
Structure of Desulfovirgula thermocuniculi IsrB (DtIsrB) in complex with omega RNA and target DNA

PDB-8dmb: 
Structure of Desulfovirgula thermocuniculi IsrB (DtIsrB) in complex with omega RNA and target DNA

EMDB-31541: 
CryoEM Structures of Reconstituted V-ATPase, Oxr1 bound V1

PDB-7fde: 
CryoEM Structures of Reconstituted V-ATPase, Oxr1 bound V1
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
