-Search query
-Search result
Showing 1 - 50 of 87 items for (author: wells & t)

EMDB-48407: 
E. coli GroEL bound with ATP and PBZ1587 inhibitor
Method: single particle / : Johnson SM, Chen Q

EMDB-48408: 
E. coli SR1 single-ring GroEL oligomer
Method: single particle / : Johnson SM, Chen Q

EMDB-48409: 
E. coli SR1 single-ring GroEL templated into pseudo-double-ring complex with PBZ1587 inhibitor
Method: single particle / : Johnson SM, Chen Q

EMDB-48410: 
E. coli SR1 single-ring GroEL oligomer
Method: single particle / : Johnson SM, Chen Q

EMDB-48411: 
E. coli GroES-GroEL-GroES football complex
Method: single particle / : Johnson SM, Chen Q

EMDB-46758: 
Cryo-EM structure of neutralizing murine antibody WS.HSV-1.24 in complex with HSV-1 glycoprotein B trimer gB-Ecto.516P.531E
Method: single particle / : Roark RS, Shapiro LS, Kwong PD

EMDB-46759: 
Cryo-EM structure of neutralizing human antibody D48 in complex with HSV-1 glycoprotein B trimer gB-Ecto.516P.531E.DS
Method: single particle / : Roark RS, Shapiro L, Kwong PD

EMDB-46760: 
Cryo-EM structure of neutralizing human antibody D48 in complex with HSV-1 glycoprotein B trimer gB-Ecto.516P
Method: single particle / : Roark RS, Shapiro L, Kwong PD

EMDB-46761: 
Cryo-EM structure of neutralizing human antibody D48 in complex with HSV-1 glycoprotein B trimer gB-Ecto.516P.531E
Method: single particle / : Roark RS, Shapiro LS, Kwong PD

EMDB-46762: 
Cryo-EM structure of gB-Ecto.516P.531E.DS, a prefusion-stabilized HSV-1 glycoprotein B extracellular domain
Method: single particle / : Roark RS, Shapiro L, Kwong PD

EMDB-46765: 
Cryo-EM structure of gB-Ecto.516P, an HSV-1 glycoprotein B extracellular domain
Method: single particle / : Roark RS, Lawrence L, Kwong PD

PDB-9dd6: 
Cryo-EM structure of neutralizing murine antibody WS.HSV-1.24 in complex with HSV-1 glycoprotein B trimer gB-Ecto.516P.531E
Method: single particle / : Roark RS, Shapiro LS, Kwong PD

PDB-9dd7: 
Cryo-EM structure of neutralizing human antibody D48 in complex with HSV-1 glycoprotein B trimer gB-Ecto.516P.531E.DS
Method: single particle / : Roark RS, Shapiro L, Kwong PD

PDB-9dd8: 
Cryo-EM structure of neutralizing human antibody D48 in complex with HSV-1 glycoprotein B trimer gB-Ecto.516P
Method: single particle / : Roark RS, Shapiro L, Kwong PD

PDB-9dd9: 
Cryo-EM structure of neutralizing human antibody D48 in complex with HSV-1 glycoprotein B trimer gB-Ecto.516P.531E
Method: single particle / : Roark RS, Shapiro LS, Kwong PD

PDB-9dda: 
Cryo-EM structure of gB-Ecto.516P.531E.DS, a prefusion-stabilized HSV-1 glycoprotein B extracellular domain
Method: single particle / : Roark RS, Shapiro L, Kwong PD

PDB-9ddc: 
Cryo-EM structure of gB-Ecto.516P, an HSV-1 glycoprotein B extracellular domain
Method: single particle / : Roark RS, Lawrence L, Kwong PD

EMDB-19566: 
Human OCCM DNA licensing intermediate
Method: single particle / : Wells JN, Leber V, Edwards LV, Allyjaun S, Peach M, Tomkins J, Kefala-Stavridi A, Faull SV, Aramayo R, Pestana CM, Ranjha L, Speck C

PDB-8rwv: 
Human OCCM DNA licensing intermediate
Method: single particle / : Wells JN, Leber V, Edwards LV, Allyjaun S, Peach M, Tomkins J, Kefala-Stavridi A, Faull SV, Aramayo R, Pestana CM, Ranjha L, Speck C
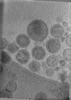
EMDB-47705: 
Tomogram of Cas9 containing Enveloped Delivery Vehicles (EDVs)
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J
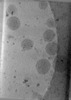
EMDB-47741: 
Tomogram of Enveloped Delivery Vehicles (EDVs) without Cas9
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-47743: 
Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) without Cas9
Method: subtomogram averaging / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-47745: 
Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) containing Cas9
Method: subtomogram averaging / : Peukes J, Ngo W, Nogales E, Doudna J
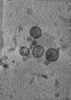
EMDB-47858: 
Tomogram of a minimized Enveloped Delivery Vehicle (miniEDV)
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-19132: 
Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Mladenov M, Seda M, Jenkins D, Stephens DJ, Roberts AJ

EMDB-19133: 
Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Seda M, Jenkins D, Stephens DJ, Roberts AJ

PDB-8rgg: 
Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Mladenov M, Seda M, Jenkins D, Stephens DJ, Roberts AJ

PDB-8rgh: 
Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Seda M, Jenkins D, Stephens DJ, Roberts AJ

EMDB-40589: 
hPAD4 bound to Activating Fab hA362
Method: single particle / : Maker A, Verba KA

EMDB-40590: 
hPAD4 bound to inhibitory Fab hI365
Method: single particle / : Maker A, Verba KA

EMDB-29395: 
Subtomogram average of HuCoV-NL63 spike protein from purified intact virions
Method: subtomogram averaging / : Chen M, Chmielewski D, Schmid M, Jin J, Chiu W

EMDB-17958: 
Structure of DPS determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17959: 
Structure of bacterial ribosome determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17960: 
Structure of GABAAR determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17961: 
Structure of mouse heavy-chain apoferritin determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17962: 
Structure of catalase determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17963: 
Structure of AHIR determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17964: 
Structure of GAPDH determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17965: 
Structure of E. coli glutamine synthetase determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17966: 
Structure of human apo ALDH1A1 determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17967: 
Structure of PaaZ determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

EMDB-17968: 
Structure of lumazine synthase determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pv9: 
Structure of DPS determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pva: 
Structure of bacterial ribosome determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pvb: 
Structure of GABAAR determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pvc: 
Structure of mouse heavy-chain apoferritin determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pvd: 
Structure of catalase determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ

PDB-8pve: 
Structure of AHIR determined by cryoEM at 100 keV
Method: single particle / : McMullan G, Naydenova K, Mihaylov D, Peet MJ, Wilson H, Yamashita K, Dickerson JL, Chen S, Cannone G, Lee Y, Hutchings KA, Gittins O, Sobhy M, Wells T, El-Gomati MM, Dalby J, Meffert M, Schulze-Briese C, Henderson R, Russo CJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
