+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of GAPDH determined by cryoEM at 100 keV | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | GAPDH / OXIDOREDUCTASE | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationglyceraldehyde-3-phosphate dehydrogenase (phosphorylating) / glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity / glycolytic process / glucose metabolic process / NAD binding / NADP binding / identical protein binding / cytosol Similarity search - Function | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
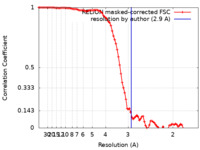
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||||||||||||||
 Authors Authors | McMullan G / Naydenova K / Mihaylov D / Peet MJ / Wilson H / Yamashita K / Dickerson JL / Chen S / Cannone G / Lee Y ...McMullan G / Naydenova K / Mihaylov D / Peet MJ / Wilson H / Yamashita K / Dickerson JL / Chen S / Cannone G / Lee Y / Hutchings KA / Gittins O / Sobhy M / Wells T / El-Gomati MM / Dalby J / Meffert M / Schulze-Briese C / Henderson R / Russo CJ | |||||||||||||||||||||
| Funding support |  United Kingdom, 6 items United Kingdom, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Structure determination by cryoEM at 100 keV. Authors: Greg McMullan / Katerina Naydenova / Daniel Mihaylov / Keitaro Yamashita / Mathew J Peet / Hugh Wilson / Joshua L Dickerson / Shaoxia Chen / Giuseppe Cannone / Yang Lee / Katherine A ...Authors: Greg McMullan / Katerina Naydenova / Daniel Mihaylov / Keitaro Yamashita / Mathew J Peet / Hugh Wilson / Joshua L Dickerson / Shaoxia Chen / Giuseppe Cannone / Yang Lee / Katherine A Hutchings / Olivia Gittins / Mohamed A Sobhy / Torquil Wells / Mohamed M El-Gomati / Jason Dalby / Matthias Meffert / Clemens Schulze-Briese / Richard Henderson / Christopher J Russo /    Abstract: Electron cryomicroscopy can, in principle, determine the structures of most biological molecules but is currently limited by access, specimen preparation difficulties, and cost. We describe a purpose- ...Electron cryomicroscopy can, in principle, determine the structures of most biological molecules but is currently limited by access, specimen preparation difficulties, and cost. We describe a purpose-built instrument operating at 100 keV-including advances in electron optics, detection, and processing-that makes structure determination fast and simple at a fraction of current costs. The instrument attains its theoretical performance limits, allowing atomic resolution imaging of gold test specimens and biological molecular structure determination in hours. We demonstrate its capabilities by determining the structures of eleven different specimens, ranging in size from 140 kDa to 2 MDa, using a fraction of the data normally required. CryoEM with a microscope designed specifically for high-efficiency, on-the-spot imaging of biological molecules will expand structural biology to a wide range of previously intractable problems. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17964.map.gz emd_17964.map.gz | 6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17964-v30.xml emd-17964-v30.xml emd-17964.xml emd-17964.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17964_fsc.xml emd_17964_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_17964.png emd_17964.png | 91.9 KB | ||
| Masks |  emd_17964_msk_1.map emd_17964_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17964.cif.gz emd-17964.cif.gz | 6.2 KB | ||
| Others |  emd_17964_half_map_1.map.gz emd_17964_half_map_1.map.gz emd_17964_half_map_2.map.gz emd_17964_half_map_2.map.gz | 48.5 MB 48.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17964 http://ftp.pdbj.org/pub/emdb/structures/EMD-17964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17964 | HTTPS FTP |
-Related structure data
| Related structure data |  8pvfMC  8pv9C  8pvaC  8pvbC  8pvcC  8pvdC  8pveC  8pvgC  8pvhC  8pviC  8pvjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17964.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17964.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8415 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17964_msk_1.map emd_17964_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17964_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17964_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Glyceraldehyde 3-phosphate dehydrogenase
| Entire | Name: Glyceraldehyde 3-phosphate dehydrogenase |
|---|---|
| Components |
|
-Supramolecule #1: Glyceraldehyde 3-phosphate dehydrogenase
| Supramolecule | Name: Glyceraldehyde 3-phosphate dehydrogenase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glyceraldehyde-3-phosphate dehydrogenase
| Macromolecule | Name: Glyceraldehyde-3-phosphate dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.059484 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHSSG RENLYFQGTL GINGFGRIGR LVLRACMERN DITVVAINDP FMDVEYMAYL LKYDSVHGNF NGTVEVSGKD LCINGKVVK VFQAKDPAEI PWGASGAQIV CESTGVFTTE EKASLHLKGG AKKVIISAPP KDNVPMYVMG VNNTEYDPSK F NVISNASC ...String: MHHHHHHSSG RENLYFQGTL GINGFGRIGR LVLRACMERN DITVVAINDP FMDVEYMAYL LKYDSVHGNF NGTVEVSGKD LCINGKVVK VFQAKDPAEI PWGASGAQIV CESTGVFTTE EKASLHLKGG AKKVIISAPP KDNVPMYVMG VNNTEYDPSK F NVISNASC TTNCLAPLAK IINDKFGIVE GLMTTVHSLT ANQLTVDGPS KGGKDWRAGR CAGNNIIPAS TGAAKAVGKV IP ALNGKLT GMAIRVPTPD VSVVDLTCKL AKPASIEEIY QAVKEASNGP MKGIMGYTSD DVVSTDFIGC KYSSIFDKNA CIA LNDSFV KLISWYDNES GYSNRLVDLA VYVASRGL UniProtKB: Glyceraldehyde-3-phosphate dehydrogenase |
-Macromolecule #2: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 1 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: UltrAuFoil R0./1 / Material: GOLD |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1400/HR + YPS FEG |
|---|---|
| Image recording | Film or detector model: DECTRIS SINGLA (1k x 1k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)