+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of GABAAR determined by cryoEM at 100 keV | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Pentameric ligand-gated ion channel / Neurotrasmitter receptor / GABA(A) receptor / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcircadian sleep/wake cycle, REM sleep / reproductive behavior / hard palate development / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex ...circadian sleep/wake cycle, REM sleep / reproductive behavior / hard palate development / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / innervation / response to anesthetic / postsynaptic specialization membrane / inhibitory postsynaptic potential / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / cellular response to zinc ion / exploration behavior / motor behavior / roof of mouth development / Signaling by ERBB4 / cochlea development / social behavior / chloride channel complex / chloride transmembrane transport / cytoplasmic vesicle membrane / cerebellum development / learning / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / GABA-ergic synapse / memory / dendritic spine / postsynaptic membrane / response to xenobiotic stimulus / cell surface / signal transduction / identical protein binding / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||||||||
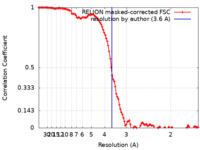
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||||||||||||||
 Authors Authors | McMullan G / Naydenova K / Mihaylov D / Peet MJ / Wilson H / Yamashita K / Dickerson JL / Chen S / Cannone G / Lee Y ...McMullan G / Naydenova K / Mihaylov D / Peet MJ / Wilson H / Yamashita K / Dickerson JL / Chen S / Cannone G / Lee Y / Hutchings KA / Gittins O / Sobhy M / Wells T / El-Gomati MM / Dalby J / Meffert M / Schulze-Briese C / Henderson R / Russo CJ | |||||||||||||||||||||
| Funding support |  United Kingdom, 6 items United Kingdom, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Structure determination by cryoEM at 100 keV. Authors: Greg McMullan / Katerina Naydenova / Daniel Mihaylov / Keitaro Yamashita / Mathew J Peet / Hugh Wilson / Joshua L Dickerson / Shaoxia Chen / Giuseppe Cannone / Yang Lee / Katherine A ...Authors: Greg McMullan / Katerina Naydenova / Daniel Mihaylov / Keitaro Yamashita / Mathew J Peet / Hugh Wilson / Joshua L Dickerson / Shaoxia Chen / Giuseppe Cannone / Yang Lee / Katherine A Hutchings / Olivia Gittins / Mohamed A Sobhy / Torquil Wells / Mohamed M El-Gomati / Jason Dalby / Matthias Meffert / Clemens Schulze-Briese / Richard Henderson / Christopher J Russo /    Abstract: Electron cryomicroscopy can, in principle, determine the structures of most biological molecules but is currently limited by access, specimen preparation difficulties, and cost. We describe a purpose- ...Electron cryomicroscopy can, in principle, determine the structures of most biological molecules but is currently limited by access, specimen preparation difficulties, and cost. We describe a purpose-built instrument operating at 100 keV-including advances in electron optics, detection, and processing-that makes structure determination fast and simple at a fraction of current costs. The instrument attains its theoretical performance limits, allowing atomic resolution imaging of gold test specimens and biological molecular structure determination in hours. We demonstrate its capabilities by determining the structures of eleven different specimens, ranging in size from 140 kDa to 2 MDa, using a fraction of the data normally required. CryoEM with a microscope designed specifically for high-efficiency, on-the-spot imaging of biological molecules will expand structural biology to a wide range of previously intractable problems. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17960.map.gz emd_17960.map.gz | 10 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17960-v30.xml emd-17960-v30.xml emd-17960.xml emd-17960.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17960_fsc.xml emd_17960_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_17960.png emd_17960.png | 106.7 KB | ||
| Masks |  emd_17960_msk_1.map emd_17960_msk_1.map | 120.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17960.cif.gz emd-17960.cif.gz | 7.4 KB | ||
| Others |  emd_17960_half_map_1.map.gz emd_17960_half_map_1.map.gz emd_17960_half_map_2.map.gz emd_17960_half_map_2.map.gz | 94.7 MB 94.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17960 http://ftp.pdbj.org/pub/emdb/structures/EMD-17960 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17960 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17960 | HTTPS FTP |
-Related structure data
| Related structure data |  8pvbMC  8pv9C  8pvaC  8pvcC  8pvdC  8pveC  8pvfC  8pvgC  8pvhC  8pviC  8pvjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17960.map.gz / Format: CCP4 / Size: 120.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17960.map.gz / Format: CCP4 / Size: 120.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
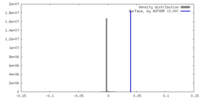
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17960_msk_1.map emd_17960_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
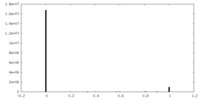
| Density Histograms |
-Half map: #1
| File | emd_17960_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
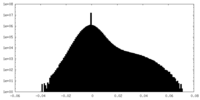
| Density Histograms |
-Half map: #2
| File | emd_17960_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
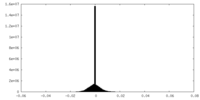
| Density Histograms |
- Sample components
Sample components
-Entire : Human GABA(A)R-beta3 homopentamer in complex with Megabody-25 in ...
| Entire | Name: Human GABA(A)R-beta3 homopentamer in complex with Megabody-25 in lipid nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: Human GABA(A)R-beta3 homopentamer in complex with Megabody-25 in ...
| Supramolecule | Name: Human GABA(A)R-beta3 homopentamer in complex with Megabody-25 in lipid nanodisc type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gamma-aminobutyric acid receptor subunit beta-3
| Macromolecule | Name: Gamma-aminobutyric acid receptor subunit beta-3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.744285 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EMDEKTTGWR GGHVVEGLAG ELEQLRARLE HHPQGQREPD YDIPTTENLY FQGTGQSVND PGNMSFVKET VDKLLKGYDI RLRPDFGGP PVCVGMNIDI ASIDMVSEVN MDYTLTMYFQ QYWRDKRLAY SGIPLNLTLD NRVADQLWVP DTYFLNDKKS F VHGVTVKN ...String: EMDEKTTGWR GGHVVEGLAG ELEQLRARLE HHPQGQREPD YDIPTTENLY FQGTGQSVND PGNMSFVKET VDKLLKGYDI RLRPDFGGP PVCVGMNIDI ASIDMVSEVN MDYTLTMYFQ QYWRDKRLAY SGIPLNLTLD NRVADQLWVP DTYFLNDKKS F VHGVTVKN RMIRLHPDGT VLYGLRITTT AACMMDLRRY PLDEQNCTLE IESYGYTTDD IEFYWRGGDK AVTGVERIEL PQ FSIVEHR LVSRNVVFAT GAYPRLSLSF RLKRNIGYFI LQTYMPSILI TILSWVSFWI NYDASAARVA LGITTVLTMT TIN THLRET LPKIPYVTAI DMYLMGCFVF VFLALLEYAF VNYIFFSQPA RAAAIDRWSR IVFPFTFSLF NLVYWLYYVN UniProtKB: Gamma-aminobutyric acid receptor subunit beta-3 |
-Macromolecule #2: Megabody Mb25
| Macromolecule | Name: Megabody Mb25 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.300629 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLVESGGG LVQTKTTTSV IDTTNDAQNL LTQAQTIVNT LKDYCPILIA KSSSSNGGTN NANTPSWQTA GGGKNSCATF GAEFSAASD MINNAQKIVQ ETQQLSANQP KNITQPHNLN LNSPSSLTAL AQKMLKNAQS QAEILKLANQ VESDFNKLSS G HLKDYIGK ...String: QVQLVESGGG LVQTKTTTSV IDTTNDAQNL LTQAQTIVNT LKDYCPILIA KSSSSNGGTN NANTPSWQTA GGGKNSCATF GAEFSAASD MINNAQKIVQ ETQQLSANQP KNITQPHNLN LNSPSSLTAL AQKMLKNAQS QAEILKLANQ VESDFNKLSS G HLKDYIGK CDASAISSAN MTMQNQKNNW GNGCAGVEET QSLLKTSAAD FNNQTPQINQ AQNLANTLIQ ELGNNTYEQL SR LLTNDNG TNSKTSAQAI NQAVNNLNER AKTLAGGTTN SPAYQATLLA LRSVLGLWNS MGYAVICGGY TKSPGENNQK DFH YTDENG NGTTINCGGS TNSNGTHSYN GTNTLKADKN VSLSIEQYEK IHEAYQILSK ALKQAGLAPL NSKGEKLEAH VTTS KYGSL RLSCAASGHT FNYPIMGWFR QAPGKEREFV GAISWSGGST SYADSVKDRF TISRDNAKNT VYLEMNNLKP EDTAV YYCA AKGRYSGGLY YPTNYDYWGQ GTQVTVSSHH HHHHEPEA |
-Macromolecule #5: HEXADECANE
| Macromolecule | Name: HEXADECANE / type: ligand / ID: 5 / Number of copies: 1 / Formula: R16 |
|---|---|
| Molecular weight | Theoretical: 226.441 Da |
| Chemical component information |  ChemComp-R16: |
-Macromolecule #6: DECANE
| Macromolecule | Name: DECANE / type: ligand / ID: 6 / Number of copies: 4 / Formula: D10 |
|---|---|
| Molecular weight | Theoretical: 142.282 Da |
| Chemical component information |  ChemComp-D10: |
-Macromolecule #7: HISTAMINE
| Macromolecule | Name: HISTAMINE / type: ligand / ID: 7 / Number of copies: 1 / Formula: HSM |
|---|---|
| Molecular weight | Theoretical: 111.145 Da |
| Chemical component information | 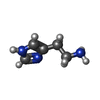 ChemComp-HSM: |
-Macromolecule #8: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 8 / Number of copies: 2 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #9: Etomidate
| Macromolecule | Name: Etomidate / type: ligand / ID: 9 / Number of copies: 1 / Formula: V8D |
|---|---|
| Molecular weight | Theoretical: 244.289 Da |
| Chemical component information | 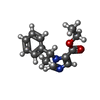 ChemComp-V8D: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: UltrAuFoil R0./1 / Material: GOLD |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1400/HR + YPS FEG |
|---|---|
| Image recording | Film or detector model: DECTRIS SINGLA (1k x 1k) / Digitization - Dimensions - Width: 1030 pixel / Digitization - Dimensions - Height: 1066 pixel / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)