-Search query
-Search result
Showing 1 - 50 of 281 items for (author: ung & kl)
EMDB-36488: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)

EMDB-37212: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Receptor original map)
EMDB-37214: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Ligand/CCL7 focused map)

PDB-8jps: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)
EMDB-43712: 
Human EBP complexed with compound 1
EMDB-43713: 
Human EBP complexed with compound 3a

PDB-8w0r: 
Human EBP complexed with compound 1

PDB-8w0s: 
Human EBP complexed with compound 3a

EMDB-19798: 
human PLD3 homodimer structure

PDB-8s86: 
human PLD3 homodimer structure

EMDB-43736: 
Umb1 umbrella toxin particle

EMDB-43737: 
Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF of UmbC1 and UmbA1)

EMDB-41963: 
Preholo-Proteasome from Beta 3 D205 deletion

EMDB-41993: 
Proteasome 20S Core Particle from Beta 3 D205 deletion

PDB-8u6y: 
Preholo-Proteasome from Beta 3 D205 deletion

PDB-8u7u: 
Proteasome 20S Core Particle from Beta 3 D205 deletion

EMDB-40799: 
Cryo-EM consensus map of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex

EMDB-40589: 
hPAD4 bound to Activating Fab hA362

EMDB-40590: 
hPAD4 bound to inhibitory Fab hI365

PDB-8smk: 
hPAD4 bound to Activating Fab hA362

PDB-8sml: 
hPAD4 bound to inhibitory Fab hI365

EMDB-36948: 
Human gamma-secretase in complex with a substrate mimetic

PDB-8k8e: 
Human gamma-secretase in complex with a substrate mimetic

EMDB-17705: 
Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class I

EMDB-17706: 
Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class II

EMDB-17707: 
Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class III

EMDB-17709: 
Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class V

EMDB-17710: 
Structure of Chelator-GIDSR4 - Fbp1 - phospho-Ubc8~ubiquitin - class IV

EMDB-17713: 
Catalytic module of human CTLH E3 ligase bound to multiphosphorylated UBE2H~ubiquitin

EMDB-17715: 
SRS and Cat modules of human CTLHSR4 bound to multiphosphorylated UBE2H~ubiquitin

EMDB-17716: 
Structure of CTLHSR4 - phospho-UBE2H~ubiquitin bound to engineered VH
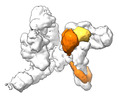
EMDB-17717: 
SRS and Cat modules of yeast Chelator-GIDSR4 bound to multiphosphorylated Ubc8~ubiquitin
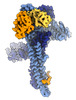
EMDB-17764: 
Catalytic module of yeast GID E3 ligase bound to multiphosphorylated Ubc8~ubiquitin

PDB-8pjn: 
Catalytic module of human CTLH E3 ligase bound to multiphosphorylated UBE2H~ubiquitin

PDB-8pmq: 
Catalytic module of yeast GID E3 ligase bound to multiphosphorylated Ubc8~ubiquitin
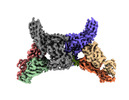
EMDB-34174: 
Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human Atypical chemokine receptor 2, ACKR2 (D6R)
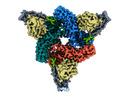
EMDB-36078: 
Structure of beta-arrestin2 in complex with M2Rpp
EMDB-36081: 
Structure of beta-arrestin2 in complex with D6Rpp (Local Refine)
EMDB-36082: 
Structure of beta-arrestin1 in complex with D6Rpp
EMDB-36090: 
Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local refine, cross-linked)
EMDB-36091: 
Muscarinic receptor (M2R) in complex with beta-arrestin1 (Low resolution full map, Cross-linked)
EMDB-36093: 
Muscarinic receptor (M2R) in complex with beta-arrestin1 (Low resolution full map, non-crosslinked)

EMDB-36110: 
Structure of basal beta-arrestin2
EMDB-36124: 
Structure of beta-arrestin1 in complex with C3aRpp
EMDB-36126: 
Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local Refine, non-cross linked)

PDB-8go9: 
Structure of beta-arrestin2 in complex with a phosphopeptide corresponding to the human Atypical chemokine receptor 2, ACKR2 (D6R)

PDB-8j8r: 
Structure of beta-arrestin2 in complex with M2Rpp

PDB-8j8v: 
Structure of beta-arrestin2 in complex with D6Rpp (Local Refine)

PDB-8j8z: 
Structure of beta-arrestin1 in complex with D6Rpp

PDB-8j97: 
Structure of Muscarinic receptor (M2R) in complex with beta-arrestin1 (Local refine, cross-linked)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
