-Search query
-Search result
Showing all 49 items for (author: stephane & r)

EMDB-44061: 
Artemia franciscana ATP synthase state 2 (composite structure), pH 7.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44087: 
Artemia franciscana ATP synthase state 1, pH 7.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44094: 
Artemia franciscana ATP synthase state 3a, pH 7.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44096: 
Artemia franciscana ATP synthase F1 domain, state 3a, pH 7.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44142: 
Artemia franciscana ATP synthase state 2 (composite structure), pH 8.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44162: 
Artemia franciscana ATP synthase FO domain, state 2, pH 7.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44165: 
Artemia franciscana ATP synthase F1 domain, state 2, pH 7.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44169: 
Artemia franciscana ATP synthase FO domain, state 2, pH 8.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44170: 
Artemia franciscana ATP synthase F1 domain, state 2, pH 8.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44172: 
Artemia franciscana ATP synthase peripheral stalk, state 2, pH 8.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44173: 
Artemia franciscana ATP synthase, state 1, pH 8.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44177: 
Artemia franciscana ATP synthase, state 2, FOF1 with weak density of the peripheral stalk, pH 8.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-44776: 
Artemia franciscana ATP synthase FO domain, state 1, pH 7.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-49579: 
Artemia franciscana ATP synthase, state 2, pH 8.0, consensus map
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-49580: 
Artemia franciscana ATP synthase, state 2, pH 7.0, consensus map
Method: single particle / : Mnatsakanyan N, Mello JFR

PDB-9b0x: 
Artemia franciscana ATP synthase state 2 (composite structure), pH 7.0
Method: single particle / : Mnatsakanyan N, Mello JFR

PDB-9b3j: 
Artemia franciscana ATP synthase state 2 (composite structure), pH 8.0
Method: single particle / : Mnatsakanyan N, Mello JFR

PDB-9bpg: 
Artemia franciscana ATP synthase FO domain, state 1, pH 7.0
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-43079: 
Artemia franciscana ATP synthase state 3a, pH 8
Method: single particle / : Mnatsakanyan N, Mello JFR

EMDB-17766: 
CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa japonica group
Method: single particle / : Huang LY, Rety S, Xi XG

EMDB-17768: 
CryoEM structure of Nal1 protein, allele IR64, from Oryza sativa indica cultivar
Method: single particle / : Huang LY, Rety S, Xi XG

PDB-8pn1: 
CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa japonica group
Method: single particle / : Huang LY, Rety S, Xi XG

PDB-8pn2: 
CryoEM structure of Nal1 protein, allele IR64, from Oryza sativa indica cultivar
Method: single particle / : Huang LY, Rety S, Xi XG

EMDB-28910: 
Glycan-Base ConC Env Trimer
Method: single particle / : Olia AS, Kwong PD

EMDB-14922: 
cryo-EM structure of omicron spike in complex with de novo designed binder, full map
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

PDB-7zrv: 
cryo-EM structure of omicron spike in complex with de novo designed binder, full map
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

EMDB-14930: 
cryo-EM structure of omicron spike in complex with de novo designed binder, local
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

EMDB-14947: 
cryo-EM structure of D614 spike in complex with de novo designed binder, full and local maps(addition)
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

PDB-7zsd: 
cryo-EM structure of omicron spike in complex with de novo designed binder, local
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

PDB-7zss: 
cryo-EM structure of D614 spike in complex with de novo designed binder
Method: single particle / : Pablo G, Sarah W, Alexandra VH, Anthony M, Andreas S, Zander H, Dongchun N, Shuguang T, Freyr S, Casper G, Priscilla T, Alexandra T, Stephane R, Sandrine G, Jane M, Aaron P, Zepeng X, Yan C, Pu H, George G, Elisa O, Beat F, Didier T, Henning S, Michael B, Bruno EC

EMDB-25794: 
Cryo-EM structure of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1
Method: single particle / : Zhou T, Tsybovsky T

EMDB-25797: 
Locally refined region of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1
Method: single particle / : Zhou T, Tsybovsky T

EMDB-25806: 
Locally refined region of SARS-CoV-2 spike in complex with antibody A19-46.1
Method: single particle / : Zhou T, Kwong PD

EMDB-25807: 
Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibody A19-46.1
Method: single particle / : Zhou T, Kwong PD

EMDB-25808: 
Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibodies A19-46.1 and B1-182.1
Method: single particle / : Zhou T, Kwong PD

EMDB-26256: 
Local refinement of cryo-EM structure of the interface of the Omicron RBD in complex with antibodies B-182.1 and A19-46.1
Method: single particle / : Zhou T, kwong PD

PDB-7tb8: 
Cryo-EM structure of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

PDB-7tbf: 
Locally refined region of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

PDB-7tc9: 
Locally refined region of SARS-CoV-2 spike in complex with antibody A19-46.1
Method: single particle / : Zhou T, Kwong PD

PDB-7tca: 
Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibody A19-46.1
Method: single particle / : Zhou T, Kwong PD

PDB-7tcc: 
Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibodies A19-46.1 and B1-182.1
Method: single particle / : Zhou T, Kwong PD

PDB-7u0d: 
Local refinement of cryo-EM structure of the interface of the Omicron RBD in complex with antibodies B-182.1 and A19-46.1
Method: single particle / : Zhou T, kwong PD

EMDB-23480: 
Cryo-EM structure of llama J3 VHH antibody in complex with HIV-1 Env BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-7lpn: 
Cryo-EM structure of llama J3 VHH antibody in complex with HIV-1 Env BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-5a42: 
Cryo-EM single particle 3D reconstruction of the native conformation of E. coli alpha-2-macroglobulin (ECAM)
Method: single particle / : Garcia-Ferrer I, Arede P, Gomez-Blanco J, Luque D, Duquerroy S, Caston JR, Goulas T, Gomis-Ruth FX

EMDB-3016: 
Cryo-EM single particle 3D reconstruction of the native conformation of E. coli alpha-2-macroglobulin (ECAM)
Method: single particle / : Garcia Ferrer I, Arede P, Gomez Blanco J, Luque D, Duquerroy S, Caston JR, Goulas T, Gomis Ruth FX
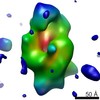
EMDB-3017: 
Cryo-EM single particle 3D reconstruction of the trypsin-induced dimeric conformation of E. coli alpha-2-macroglobulin (ECAM)
Method: single particle / : Garcia Ferrer I, Arede P, Gomez Blanco J, Luque D, Duquerroy S, Caston JR, Goulas T, Gomis Ruth FX

EMDB-3018: 
Cryo-EM single particle 3D reconstruction of the induced, monomeric conformation of E. coli alpha-2-macroglobulin (ECAM)
Method: single particle / : Garcia Ferrer I, Arede P, Gomez Blanco J, Luque D, Duquerroy S, Caston JR, Goulas T, Gomis Ruth FX
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
