[English] 日本語
 Yorodumi
Yorodumi- EMDB-3018: Cryo-EM single particle 3D reconstruction of the induced, monomer... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3018 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM single particle 3D reconstruction of the induced, monomeric conformation of E. coli alpha-2-macroglobulin (ECAM) | |||||||||
 Map data Map data | Reconstruction of induced monomeric Escherichia coli alpha-2-macroglobulin | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | alpha-2-macroglobulin / ECAM / peptidase inhibitor | |||||||||
| Function / homology |  Function and homology information Function and homology informationendopeptidase inhibitor activity / defense response / protein homodimerization activity / extracellular space / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 17.0 Å | |||||||||
 Authors Authors | Garcia Ferrer I / Arede P / Gomez Blanco J / Luque D / Duquerroy S / Caston JR / Goulas T / Gomis Ruth FX | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2015 Journal: Proc Natl Acad Sci U S A / Year: 2015Title: Structural and functional insights into Escherichia coli α2-macroglobulin endopeptidase snap-trap inhibition. Authors: Irene Garcia-Ferrer / Pedro Arêde / Josué Gómez-Blanco / Daniel Luque / Stephane Duquerroy / José R Castón / Theodoros Goulas / F Xavier Gomis-Rüth /   Abstract: The survival of commensal bacteria requires them to evade host peptidases. Gram-negative bacteria from the human gut microbiome encode a relative of the human endopeptidase inhibitor, α2- ...The survival of commensal bacteria requires them to evade host peptidases. Gram-negative bacteria from the human gut microbiome encode a relative of the human endopeptidase inhibitor, α2-macroglobulin (α2M). Escherichia coli α2M (ECAM) is a ∼ 180-kDa multidomain membrane-anchored pan-peptidase inhibitor, which is cleaved by host endopeptidases in an accessible bait region. Structural studies by electron microscopy and crystallography reveal that this cleavage causes major structural rearrangement of more than half the 13-domain structure from a native to a compact induced form. It also exposes a reactive thioester bond, which covalently traps the peptidase. Subsequently, peptidase-laden ECAM is shed from the membrane and may dimerize. Trapped peptidases are still active except against very large substrates, so inhibition potentially prevents damage of large cell envelope components, but not host digestion. Mechanistically, these results document a novel monomeric "snap trap." | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3018.map.gz emd_3018.map.gz | 1.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3018-v30.xml emd-3018-v30.xml emd-3018.xml emd-3018.xml | 9.1 KB 9.1 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-3018image500gris.png EMD-3018image500gris.png | 34.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3018 http://ftp.pdbj.org/pub/emdb/structures/EMD-3018 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3018 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3018 | HTTPS FTP |
-Related structure data
| Related structure data |  3016C 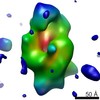 3017C  4ziqC  4ziuC  4zjgC  4zjhC  5a42C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3018.map.gz / Format: CCP4 / Size: 1.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3018.map.gz / Format: CCP4 / Size: 1.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of induced monomeric Escherichia coli alpha-2-macroglobulin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Induced, monomeric conformation of the E. coli alpha-2-macroglobu...
| Entire | Name: Induced, monomeric conformation of the E. coli alpha-2-macroglobulin (ECAM). |
|---|---|
| Components |
|
-Supramolecule #1000: Induced, monomeric conformation of the E. coli alpha-2-macroglobu...
| Supramolecule | Name: Induced, monomeric conformation of the E. coli alpha-2-macroglobulin (ECAM). type: sample / ID: 1000 / Oligomeric state: Monomeric / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 180 KDa / Theoretical: 180 KDa / Method: Size exclusion chromatography. |
-Macromolecule #1: alpha-2-macroglobulin
| Macromolecule | Name: alpha-2-macroglobulin / type: protein_or_peptide / ID: 1 / Name.synonym: ECAM Details: ECAM was reacted with trypsin and the resulting induced, monomeric ECAM was purified by size-exclusion chromatography. Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 180 KDa / Theoretical: 180 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Alpha-2-macroglobulin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 10mM Tris-HCL, 75mM NaCl |
| Grid | Details: Glow discharged carbon coated Cu/Rh 300 mesh Quantifoil R 1.2/1.3um grids. |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC Method: Blot for 1min before plunging with 5ul purified sample. |
- Electron microscopy
Electron microscopy
| Microscope | OTHER |
|---|---|
| Date | Jul 17, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (4k x 4k) / Number real images: 84 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 84269 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | The XMIPP automatic picking routine was used to select particles. |
|---|---|
| CTF correction | Details: Each image |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 17.0 Å / Resolution method: OTHER / Software - Name: XMIPP3, EMAN2 / Number images used: 18346 |
| Final two d classification | Number classes: 64 |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















