[English] 日本語
 Yorodumi
Yorodumi- EMDB-25797: Locally refined region of SARS-CoV-2 spike in complex with antibo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Locally refined region of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | |||||||||
 Map data Map data | Locally refined cryo-EM map of SARS-CoV-2 RBD in complex with antibody B1-182.1 and A19-61.1 FV regions, needs to be z-flipped for refinement. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / spike / antibody / VIRAL PROTEIN / VIRAL PROTEIN-Immune System complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Zhou T / Tsybovsky T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Authors: Tongqing Zhou / Lingshu Wang / John Misasi / Amarendra Pegu / Yi Zhang / Darcy R Harris / Adam S Olia / Chloe Adrienna Talana / Eun Sung Yang / Man Chen / Misook Choe / Wei Shi / I-Ting Teng ...Authors: Tongqing Zhou / Lingshu Wang / John Misasi / Amarendra Pegu / Yi Zhang / Darcy R Harris / Adam S Olia / Chloe Adrienna Talana / Eun Sung Yang / Man Chen / Misook Choe / Wei Shi / I-Ting Teng / Adrian Creanga / Claudia Jenkins / Kwanyee Leung / Tracy Liu / Erik-Stephane D Stancofski / Tyler Stephens / Baoshan Zhang / Yaroslav Tsybovsky / Barney S Graham / John R Mascola / Nancy J Sullivan / Peter D Kwong /  Abstract: The rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) B.1.1.529 (Omicron) variant and its resistance to neutralization by vaccinee and convalescent sera are driving a ...The rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) B.1.1.529 (Omicron) variant and its resistance to neutralization by vaccinee and convalescent sera are driving a search for monoclonal antibodies with potent neutralization. To provide insight into effective neutralization, we determined cryo-electron microscopy structures and evaluated receptor binding domain (RBD) antibodies for their ability to bind and neutralize B.1.1.529. Mutations altered 16% of the B.1.1.529 RBD surface, clustered on an RBD ridge overlapping the angiotensin-converting enzyme 2 (ACE2)-binding surface and reduced binding of most antibodies. Substantial inhibitory activity was retained by select monoclonal antibodies-including A23-58.1, B1-182.1, COV2-2196, S2E12, A19-46.1, S309, and LY-CoV1404-that accommodated these changes and neutralized B.1.1.529. We identified combinations of antibodies with synergistic neutralization. The analysis revealed structural mechanisms for maintenance of potent neutralization against emerging variants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25797.map.gz emd_25797.map.gz | 267.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25797-v30.xml emd-25797-v30.xml emd-25797.xml emd-25797.xml | 24.2 KB 24.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25797.png emd_25797.png | 50 KB | ||
| Masks |  emd_25797_msk_1.map emd_25797_msk_1.map | 536.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25797.cif.gz emd-25797.cif.gz | 6.9 KB | ||
| Others |  emd_25797_additional_1.map.gz emd_25797_additional_1.map.gz emd_25797_half_map_1.map.gz emd_25797_half_map_1.map.gz emd_25797_half_map_2.map.gz emd_25797_half_map_2.map.gz | 506.2 MB 497.5 MB 497.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25797 http://ftp.pdbj.org/pub/emdb/structures/EMD-25797 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25797 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25797 | HTTPS FTP |
-Related structure data
| Related structure data |  7tbfMC  7tb8C  7tc9C  7tcaC  7tccC  7u0dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25797.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25797.map.gz / Format: CCP4 / Size: 536.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally refined cryo-EM map of SARS-CoV-2 RBD in complex with antibody B1-182.1 and A19-61.1 FV regions, needs to be z-flipped for refinement. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.873 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25797_msk_1.map emd_25797_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
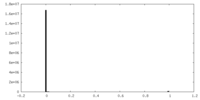
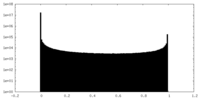
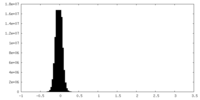
| Density Histograms |
-Additional map: Locally refined and sharpened cryo-EM map of SARS-CoV-2...
| File | emd_25797_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally refined and sharpened cryo-EM map of SARS-CoV-2 RBD in complex with antibody B1-182.1 and A19-61.1 FV regions | ||||||||||||
| Projections & Slices |
| ||||||||||||
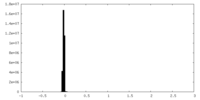
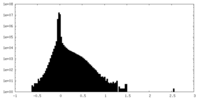
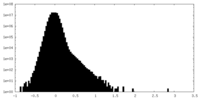
| Density Histograms |
-Half map: cryo-EM half map of SARS-CoV-2 RBD in complex...
| File | emd_25797_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM half map of SARS-CoV-2 RBD in complex with antibody B1-182.1 and A19-61.1 FV regions. | ||||||||||||
| Projections & Slices |
| ||||||||||||
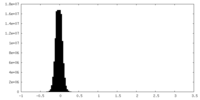
| Density Histograms |
-Half map: cryo-EM half map of SARS-CoV-2 RBD in complex...
| File | emd_25797_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM half map of SARS-CoV-2 RBD in complex with antibody B1-182.1 and A19-61.1 FV regions. | ||||||||||||
| Projections & Slices |
| ||||||||||||
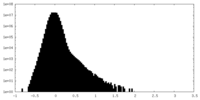
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1
| Entire | Name: SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1
| Supramolecule | Name: SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 570 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.986652 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ITNLCPFGEV FNATRFASVY AWNRKRISNC VADYSVLYNS ASFSTFKCYG VSPTKLNDLC FTNVYADSFV IRGDEVRQIA PGQTGKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF P LQSYGFQP ...String: ITNLCPFGEV FNATRFASVY AWNRKRISNC VADYSVLYNS ASFSTFKCYG VSPTKLNDLC FTNVYADSFV IRGDEVRQIA PGQTGKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF P LQSYGFQP TNGVGYQPYR VVVLSFELLH APATVCGP UniProtKB: Spike glycoprotein |
-Macromolecule #2: Heavy chain of SARS-CoV-2 antibody B1-182.1
| Macromolecule | Name: Heavy chain of SARS-CoV-2 antibody B1-182.1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.285312 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QMQLVQSGPE VKKPGTSVKV SCKASGFTFT SSAVQWVRQA RGQRLEWIGW IVVGSGNTNY AQKFQERVTI TRDMSTSTAY MELSSLRSE DTAVYYCAAP YCSGGSCFDG FDIWGQGTMV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: QMQLVQSGPE VKKPGTSVKV SCKASGFTFT SSAVQWVRQA RGQRLEWIGW IVVGSGNTNY AQKFQERVTI TRDMSTSTAY MELSSLRSE DTAVYYCAAP YCSGGSCFDG FDIWGQGTMV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSCDK |
-Macromolecule #3: Light chain of SARS-CoV-2 antibody B1-182.1
| Macromolecule | Name: Light chain of SARS-CoV-2 antibody B1-182.1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.536008 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQSVS SSYLAWYQQK PGQAPRLLIY GASSRATGFP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQYGNSPWTF GQGTKVEIRR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: EIVLTQSPGT LSLSPGERAT LSCRASQSVS SSYLAWYQQK PGQAPRLLIY GASSRATGFP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQYGNSPWTF GQGTKVEIRR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: Heavy chain of SARS-CoV-2 antibody A19-61.1
| Macromolecule | Name: Heavy chain of SARS-CoV-2 antibody A19-61.1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.934431 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVESGGG VVQPGRSLRL SCAASGFTFS SYAFHWVRQA PGKGLEWVAV ISYDGSNQYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAD DTAVYYCARD LAIAVAGTWH YYNGMDVWGQ GTTVTVSS |
-Macromolecule #5: Light chain of SARS-CoV-2 antibody A19-61.1
| Macromolecule | Name: Light chain of SARS-CoV-2 antibody A19-61.1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.571899 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS VSASVGDRVI ITCRASQGIS SWLAWYQQKP GKAPKVLIYD ASSLQSGVPS RFSGSGYGTD FTLTISSLQP EDSATYYCQ QAKSFPITFG QGTRLEIK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 100 mM HEPES, 150 mM NaCl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 2-3.5 seconds before plugging.. |
| Details | Complex at 0.5 mg/mL concentration in the buffer |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: Details: SARS-CoV-2 receptor-binding domain in complex with antibody A23-58.1 |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: cryoSPARC (ver. 2.15) / Details: Local refinement with cryoSparc / Number images used: 358526 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)