[English] 日本語
 Yorodumi
Yorodumi- EMDB-25807: Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with ant... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibody A19-46.1 | |||||||||
 Map data Map data | cryo-EM map for SARS-CoV2 Omicron spike in complex with antibody A19-46.1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / spike / antibody / VIRAL PROTEIN / VIRAL PROTEIN-Immune System complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.85 Å | |||||||||
 Authors Authors | Zhou T / Kwong PD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Authors: Tongqing Zhou / Lingshu Wang / John Misasi / Amarendra Pegu / Yi Zhang / Darcy R Harris / Adam S Olia / Chloe Adrienna Talana / Eun Sung Yang / Man Chen / Misook Choe / Wei Shi / I-Ting Teng ...Authors: Tongqing Zhou / Lingshu Wang / John Misasi / Amarendra Pegu / Yi Zhang / Darcy R Harris / Adam S Olia / Chloe Adrienna Talana / Eun Sung Yang / Man Chen / Misook Choe / Wei Shi / I-Ting Teng / Adrian Creanga / Claudia Jenkins / Kwanyee Leung / Tracy Liu / Erik-Stephane D Stancofski / Tyler Stephens / Baoshan Zhang / Yaroslav Tsybovsky / Barney S Graham / John R Mascola / Nancy J Sullivan / Peter D Kwong /  Abstract: The rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) B.1.1.529 (Omicron) variant and its resistance to neutralization by vaccinee and convalescent sera are driving a ...The rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) B.1.1.529 (Omicron) variant and its resistance to neutralization by vaccinee and convalescent sera are driving a search for monoclonal antibodies with potent neutralization. To provide insight into effective neutralization, we determined cryo-electron microscopy structures and evaluated receptor binding domain (RBD) antibodies for their ability to bind and neutralize B.1.1.529. Mutations altered 16% of the B.1.1.529 RBD surface, clustered on an RBD ridge overlapping the angiotensin-converting enzyme 2 (ACE2)-binding surface and reduced binding of most antibodies. Substantial inhibitory activity was retained by select monoclonal antibodies-including A23-58.1, B1-182.1, COV2-2196, S2E12, A19-46.1, S309, and LY-CoV1404-that accommodated these changes and neutralized B.1.1.529. We identified combinations of antibodies with synergistic neutralization. The analysis revealed structural mechanisms for maintenance of potent neutralization against emerging variants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25807.map.gz emd_25807.map.gz | 236.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25807-v30.xml emd-25807-v30.xml emd-25807.xml emd-25807.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25807.png emd_25807.png | 77.9 KB | ||
| Masks |  emd_25807_msk_1.map emd_25807_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25807.cif.gz emd-25807.cif.gz | 7.5 KB | ||
| Others |  emd_25807_additional_1.map.gz emd_25807_additional_1.map.gz emd_25807_half_map_1.map.gz emd_25807_half_map_1.map.gz emd_25807_half_map_2.map.gz emd_25807_half_map_2.map.gz | 450.8 MB 442.5 MB 442.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25807 http://ftp.pdbj.org/pub/emdb/structures/EMD-25807 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25807 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25807 | HTTPS FTP |
-Related structure data
| Related structure data |  7tcaMC  7tb8C  7tbfC  7tc9C  7tccC  7u0dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25807.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25807.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM map for SARS-CoV2 Omicron spike in complex with antibody A19-46.1 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25807_msk_1.map emd_25807_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
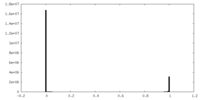
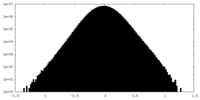
| Density Histograms |
-Additional map: Sharpened cryo-EM map for SARS-CoV2 Omicron spike in...
| File | emd_25807_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened cryo-EM map for SARS-CoV2 Omicron spike in complex with antibody A19-46.1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
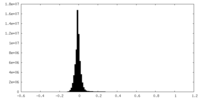
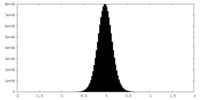
| Density Histograms |
-Half map: cryo-EM half map for SARS-CoV2 Omicron spike in...
| File | emd_25807_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM half map for SARS-CoV2 Omicron spike in complex with antibody A19-46.1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
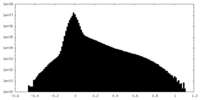
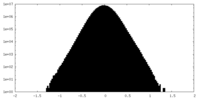
| Density Histograms |
-Half map: cryo-EM half map for SARS-CoV2 Omicron spike in...
| File | emd_25807_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM half map for SARS-CoV2 Omicron spike in complex with antibody A19-46.1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
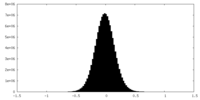
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike in complex with antibody A19-46.1
| Entire | Name: SARS-CoV-2 spike in complex with antibody A19-46.1 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike in complex with antibody A19-46.1
| Supramolecule | Name: SARS-CoV-2 spike in complex with antibody A19-46.1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 570 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 141.111234 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHVISGTNG TKRFDNPVLP FNDGVYFASI EKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLDHK NNKSWMESEF RVYSSANNCT FEYVSQPFLM D LEGKQGNF ...String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHVISGTNG TKRFDNPVLP FNDGVYFASI EKSNIIRGW IFGTTLDSKT QSLLIVNNAT NVVIKVCEFQ FCNDPFLDHK NNKSWMESEF RVYSSANNCT FEYVSQPFLM D LEGKQGNF KNLREFVFKN IDGYFKIYSK HTPIIVREPE DLPQGFSALE PLVDLPIGIN ITRFQTLLAL HRSYLTPGDS SS GWTAGAA AYYVGYLQPR TFLLKYNENG TITDAVDCAL DPLSETKCTL KSFTVEKGIY QTSNFRVQPT ESIVRFPNIT NLC PFDEVF NATRFASVYA WNRKRISNCV ADYSVLYNLA PFFTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTG NIADY NYKLPDDFTG CVIAWNSNKL DSKVSGNYNY LYRLFRKSNL KPFERDISTE IYQAGNKPCN GVAGFNCYFP LRSYS FRPT YGVGHQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLKGTG VLTESNKKFL PFQQFGRDIA DTTDAV RDP QTLEILDITP CSFGGVSVIT PGTNTSNQVA VLYQGVNCTE VPVAIHADQL TPTWRVYSTG SNVFQTRAGC LIGAEYV NN SYECDIPIGA GICASYQTQT KSHGSASSVA SQSIIAYTMS LGAENSVAYS NNSIAIPTNF TISVTTEILP VSMTKTSV D CTMYICGDST ECSNLLLQYG SFCTQLKRAL TGIAVEQDKN TQEVFAQVKQ IYKTPPIKYF GGFNFSQILP DPSKPSKRS FIEDLLFNKV TLADAGFIKQ YGDCLGDIAA RDLICAQKFK GLTVLPPLLT DEMIAQYTSA LLAGTITSGW TFGAGAALQI PFAMQMAYR FNGIGVTQNV LYENQKLIAN QFNSAIGKIQ DSLSSTASAL GKLQDVVNHN AQALNTLVKQ LSSKFGAISS V LNDIFSRL DPPEAEVQID RLITGRLQSL QTYVTQQLIR AAEIRASANL AATKMSECVL GQSKRVDFCG KGYHLMSFPQ SA PHGVVFL HVTYVPAQEK NFTTAPAICH DGKAHFPREG VFVSNGTHWF VTQRNFYEPQ IITTDNTFVS GNCDVVIGIV NNT VYDPLQ PELDSFKEEL DKYFKNHTSP DVDLGDISGI NASVVNIQKE IDRLNEVAKN LNESLIDLQE LGKYEQGSGY IPEA PRDGQ AYVRKDGEWV LLSTFLGRSL EVLFQGPGHH HHHHHHSAWS HPQFEKGGGS GGGGSGGSAW SHPQFEK UniProtKB: Spike glycoprotein |
-Macromolecule #2: Heavy chain of antibody A19-46.1
| Macromolecule | Name: Heavy chain of antibody A19-46.1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.8699 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVESGGG VVQPGRSLRL SCAASGFTLS SYGMHWVRQA PGKGLEWVAV ISYDGSNKYY VDSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCARG WAYWELLPDY YYGMDVWGQG TTVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN ...String: QVQLVESGGG VVQPGRSLRL SCAASGFTLS SYGMHWVRQA PGKGLEWVAV ISYDGSNKYY VDSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCARG WAYWELLPDY YYGMDVWGQG TTVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS CDK |
-Macromolecule #3: Light chain of antibody A19-46.1
| Macromolecule | Name: Light chain of antibody A19-46.1 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.760273 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QTVVTQEPSF SVSPGGTVTL TCGLSSGSVS TAYFPSWYQQ TPGQAPRTLI YGTNTRSSGV PDRFSGSILG NKAALTITGA QADDESDYY CVLYMGRGIV VFGGGTKLTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP ...String: QTVVTQEPSF SVSPGGTVTL TCGLSSGSVS TAYFPSWYQQ TPGQAPRTLI YGTNTRSSGV PDRFSGSILG NKAALTITGA QADDESDYY CVLYMGRGIV VFGGGTKLTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP SKQSNNKYAA SSYLSLTPEQ WKSHRSYSCQ VTHEGSTVEK TVAPTECS |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 28 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 100 mM HEPES, 150 mM NaCl |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 2-3.5 seconds before plugging.. |
| Details | Complex at 0.5 mg/mL concentration in the buffer |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: Details: SARS-CoV-2 receptor-binding domain in complex with antibody A23-58.1 |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.85 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: cryoSPARC (ver. 2.15) / Number images used: 79077 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)