[English] 日本語
 Yorodumi
Yorodumi- EMDB-23480: Cryo-EM structure of llama J3 VHH antibody in complex with HIV-1 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23480 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of llama J3 VHH antibody in complex with HIV-1 Env BG505 DS-SOSIP.664 | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CD4 / HIV-1 / SOSIP / Vaccine / IMMUNE SYSTEM / llama / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.61 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Structural basis for llama nanobody recognition and neutralization of HIV-1 at the CD4-binding site. Authors: Tongqing Zhou / Lei Chen / Jason Gorman / Shuishu Wang / Young D Kwon / Bob C Lin / Mark K Louder / Reda Rawi / Erik-Stephane D Stancofski / Yongping Yang / Baoshan Zhang / Anna Forsman ...Authors: Tongqing Zhou / Lei Chen / Jason Gorman / Shuishu Wang / Young D Kwon / Bob C Lin / Mark K Louder / Reda Rawi / Erik-Stephane D Stancofski / Yongping Yang / Baoshan Zhang / Anna Forsman Quigley / Laura E McCoy / Lucy Rutten / Theo Verrips / Robin A Weiss / / Nicole A Doria-Rose / Lawrence Shapiro / Peter D Kwong /    Abstract: Nanobodies can achieve remarkable neutralization of genetically diverse pathogens, including HIV-1. To gain insight into their recognition, we determined crystal structures of four llama nanobodies ...Nanobodies can achieve remarkable neutralization of genetically diverse pathogens, including HIV-1. To gain insight into their recognition, we determined crystal structures of four llama nanobodies (J3, A12, C8, and D7), all of which targeted the CD4-binding site, in complex with the HIV-1 envelope (Env) gp120 core, and determined a cryoelectron microscopy (cryo-EM) structure of J3 with the Env trimer. Crystal and cryo-EM structures of J3 complexes revealed this nanobody to mimic binding to the prefusion-closed trimer for the primary site of CD4 recognition as well as a secondary quaternary site. In contrast, crystal structures of A12, C8, and D7 with gp120 revealed epitopes that included portions of the gp120 inner domain, inaccessible on the prefusion-closed trimer. Overall, these structures explain the broad and potent neutralization of J3 and limited neutralization of A12, C8, and D7, which utilized binding modes incompatible with the neutralization-targeted prefusion-closed conformation of Env. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23480.map.gz emd_23480.map.gz | 79 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23480-v30.xml emd-23480-v30.xml emd-23480.xml emd-23480.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23480.png emd_23480.png | 96.2 KB | ||
| Masks |  emd_23480_msk_1.map emd_23480_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23480.cif.gz emd-23480.cif.gz | 6.7 KB | ||
| Others |  emd_23480_additional_1.map.gz emd_23480_additional_1.map.gz emd_23480_half_map_1.map.gz emd_23480_half_map_1.map.gz emd_23480_half_map_2.map.gz emd_23480_half_map_2.map.gz | 42 MB 77.8 MB 77.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23480 http://ftp.pdbj.org/pub/emdb/structures/EMD-23480 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23480 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23480 | HTTPS FTP |
-Related structure data
| Related structure data |  7lpnMC  7r73C  7r74C  7ri1C  7ri2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23480.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23480.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0605 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23480_msk_1.map emd_23480_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
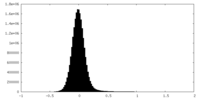
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_23480_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
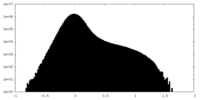
| Density Histograms |
-Half map: half map B
| File | emd_23480_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
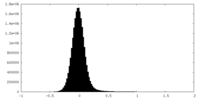
| Density Histograms |
-Half map: Half map A
| File | emd_23480_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
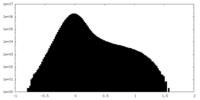
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 Env with llama J3 VHH
| Entire | Name: HIV-1 Env with llama J3 VHH |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 Env with llama J3 VHH
| Supramolecule | Name: HIV-1 Env with llama J3 VHH / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: HIV-1 Envelope Glycoprotein BG505 SOSIP.664 gp41
| Macromolecule | Name: HIV-1 Envelope Glycoprotein BG505 SOSIP.664 gp41 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp160
| Macromolecule | Name: Envelope glycoprotein gp160 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 53.300348 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SACTQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV MIRSENI TNNAKNILVQ FNTPVQINCT RPNNNTRKSI RIGPGQAFYA TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFANSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSITLPC RIKQIINMWQ RIGQ CMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG R UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: J3 VHH
| Macromolecule | Name: J3 VHH / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.489186 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVQAGGFLRL SCELRGSIFN QYAMAWFRQA PGKEREFVAG MGAVPHYGEF VKGRFTISRD NAKSTVYLQM SSLKPEDTA IYFCARSKST YISYNSNGYD YWGRGTQVTV SSAAAHHHHH H |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 33 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Formula: PBS |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 77.72 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)