-Search query
-Search result
Showing 1 - 50 of 86 items for (author: shangyu & d)

EMDB-38313: 
Structure of yeast replisome associated with FACT and histone hexamer, the region of FACT-Histones optimized local map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38314: 
Structure of yeast replisome associated with FACT and histone hexamer,Conformation-2
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38315: 
Structure of yeast replisome associated with FACT and histone hexamer, the region of polymerase epsilon optimized local map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38316: 
Structure of yeast replisome associated with FACT and histone hexamer, Conformation-1
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38317: 
Structure of yeast replisome associated with FACT and histone hexamer, Composite map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

PDB-8xgc: 
Structure of yeast replisome associated with FACT and histone hexamer, Composite map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-36313: 
Cryo-EM structure of apoferritin with MSBP
Method: single particle / : Xu Y, Qin Y, Wang L, Zhang Y, Wang Y, Dang S

EMDB-36314: 
Cryo-EM structure of hemagglutinin with MSBP
Method: single particle / : Xu Y, Qin Y, Wang L, Zhang Y, Wang Y, Dang S

EMDB-36315: 
Cryo-EM structure of catalase with MSBP
Method: single particle / : Xu Y, Qin Y, Wang L, Zhang Y, Wang Y, Dang S

EMDB-36316: 
Cryo-EM structure of beta-Galactosidase with MSBP
Method: single particle / : Xu Y, Qin Y, Wang L, Zhang Y, Wang Y, Dang S

EMDB-37345: 
Yeast replisome in state IV
Method: single particle / : Dang S, Zhai Y, Feng J, Yu D
EMDB-36759: 
Cryo-EM structure of TMEM63C
Method: single particle / : Qin Y, Yu D, Dong J, Dang S

EMDB-37211: 
Yeast replisome in state I
Method: single particle / : Dang S, Zhai Y, Feng J, Yu D, Xu Z

EMDB-37213: 
Yeast replisome in state II
Method: single particle / : Dang S, Zhai Y, Feng J, Yu D, Xu Z

EMDB-37215: 
Yeast replisome in state III
Method: single particle / : Dang S, Zhai Y, Feng J, Yu D, Xu Z

EMDB-37343: 
Yeast replisome in state V
Method: single particle / : Dang S, Zhai Y, Feng J, Yu D, Xu Z

PDB-8kg9: 
Yeast replisome in state III
Method: single particle / : Dang S, Zhai Y, Feng J, Yu D, Xu Z

EMDB-35175: 
Portal-tail complex structure of the Cyanophage P-SCSP1u
Method: single particle / : Liu H, Dang S

PDB-8i4m: 
Portal-tail complex structure of the Cyanophage P-SCSP1u
Method: single particle / : Liu H, Dang S

EMDB-35174: 
Capsid structure of the Cyanophage P-SCSP1u
Method: single particle / : Liu H, Dang S

EMDB-35074: 
Cryo-EM structure of MPXV M2 in complex with human B7.1
Method: single particle / : Wang Y, Yang S, Zhao H, Deng Z

EMDB-35075: 
Cryo-EM structure of MPXV M2 hexamer in complex with human B7.2
Method: single particle / : Wang Y, Yang S, Zhao H, Deng Z

EMDB-35076: 
Cryo-EM structure of MPXV M2 heptamer in complex with human B7.2
Method: single particle / : Wang Y, Yang S, Zhao H, Deng Z

PDB-8hxa: 
Cryo-EM structure of MPXV M2 in complex with human B7.1
Method: single particle / : Wang Y, Yang S, Zhao H, Deng Z

PDB-8hxb: 
Cryo-EM structure of MPXV M2 hexamer in complex with human B7.2
Method: single particle / : Wang Y, Yang S, Zhao H, Deng Z

PDB-8hxc: 
Cryo-EM structure of MPXV M2 heptamer in complex with human B7.2
Method: single particle / : Wang Y, Yang S, Zhao H, Deng Z

EMDB-34195: 
Human menin in complex with H3K79Me2 nucleosome
Method: single particle / : Lin J, Yu D, Lam WH, Dang S, Zhai Y, Li XD

PDB-8gpn: 
Human menin in complex with H3K79Me2 nucleosome
Method: single particle / : Lin J, Yu D, Lam WH, Dang S, Zhai Y, Li XD

EMDB-32258: 
Human MCM double hexamer bound to natural DNA duplex (polyAT/polyTA)
Method: single particle / : Li J, Dong J, Dang S, Zhai Y

EMDB-33320: 
Cryo-EM map of hMCM-DH R195A/L209G mutant
Method: single particle / : Li J, Dong JQ, Dang SY, Zhai YL

PDB-7w1y: 
Human MCM double hexamer bound to natural DNA duplex (polyAT/polyTA)
Method: single particle / : Li J, Dong J, Dang S, Zhai Y

EMDB-33892: 
Omicron spike trimer bound with P3E6 (Local Refinement)
Method: single particle / : Tang B, Dang S
EMDB-33893: 
Omicron Spike trimer bound with up P3E6 Fab (2 RBDs up and 1 RBD down)
Method: single particle / : Tang B, Dang S
EMDB-33894: 
Omicron Spike trimer bound with side P3E6 Fab (1 RBD up and 2 RBDs down)
Method: single particle / : Tang B, Dang S

PDB-7ykj: 
Omicron RBDs bound with P3E6 Fab (one up and one down)
Method: single particle / : Tang B, Dang S

EMDB-27775: 
Cryo-EM structure of RBD-directed neutralizing antibody P2B4 in complex with prefusion SARS-CoV-2 spike glycoprotein
Method: single particle / : Reddem ER, Shapiro L

PDB-8dxs: 
Cryo-EM structure of RBD-directed neutralizing antibody P2B4 in complex with prefusion SARS-CoV-2 spike glycoprotein
Method: single particle / : Reddem ER, Shapiro L
EMDB-33191: 
The structure of ZCB11 Fab against SARS-CoV-2 Omicron Spike
Method: single particle / : Hang L, Shangyu D
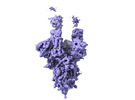
EMDB-33194: 
The Cryo-EM structure of SARS-CoV-2 Omicron 2-RBD up spike protein complexed with ZCB11 fab
Method: single particle / : Hang L, Dang S
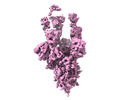
EMDB-33195: 
The Cryo-EM structure of SARS-CoV-2 Omicron 3-RBD up spike protein complexed with ZCB11 fab
Method: single particle / : Hang L, Dang S

PDB-7xh8: 
The structure of ZCB11 Fab against SARS-CoV-2 Omicron Spike
Method: single particle / : Hang L, Dang S

EMDB-31684: 
Cryo-EM structure of MCM double hexamer with structured Mcm4-NSD
Method: single particle / : Cheng J, Li N, Huo Y, Dang S, Tye B, Gao N, Zhai Y

EMDB-31685: 
Cryo-EM structure of MCM double hexamer bound with DDK in State I
Method: single particle / : Cheng J, Li N, Huo Y, Dang S, Tye B, Gao N, Zhai Y

EMDB-31686: 
Cryo-EM map of DDK subtracted from DH-DDK complex
Method: single particle / : Cheng J, Li N, Huo Y, Dang S, Tye B, Gao N, Zhai Y
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search









 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
