-Search query
-Search result
Showing all 30 items for (author: renault & l)

EMDB-14956: 
Structure of an Escherichia coli 70S ribosome stalled by Tetracenomycin X during translation of an MAAAPQK(C) peptide
Method: single particle / : Leroy EC, Perry TN, Renault TT, Innis CA

PDB-7zta: 
Structure of an Escherichia coli 70S ribosome stalled by Tetracenomycin X during translation of an MAAAPQK(C) peptide
Method: single particle / : Leroy EC, Perry TN, Renault TT, Innis CA

EMDB-13738: 
Horse spleen apoferritin
Method: single particle / : Koning RI, Renault L

EMDB-13364: 
UVC treated Human apoferritin
Method: single particle / : Renault L, Depelteau JS

EMDB-13402: 
Cryo-EM map of UVC-treated ICP1 Bacteriophage capsid
Method: single particle / : Depelteau JS, Briegel A

EMDB-13403: 
Cryo-EM map of WT ICP1 bacteriophage capsid
Method: single particle / : Depelteau JS, Briegel A

PDB-7pf1: 
UVC treated Human apoferritin
Method: single particle / : Renault L, Depelteau JS, Briegel A

EMDB-11639: 
Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(S-closed trimer)
Method: single particle / : Renault LLR, Rutten L, Juraszek J, Langedijk JPM

EMDB-11719: 
Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(One up trimer)
Method: single particle / : Renault LLR, Rutten L, Juraszek J, Langedijk JPM

PDB-7a4n: 
Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(S-closed trimer)
Method: single particle / : Rutten L, Renault LLR, Juraszek J, Langedijk JPM

PDB-7ad1: 
Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(One up trimer)
Method: single particle / : Rutten L, Renault LLR, Juraszek J, Langedijk JPM

EMDB-4692: 
Human-D02 Nucleosome Core Particle with biotin-streptavidin label
Method: single particle / : Wilson MD, Nans A, Pye VE, Cherepanov P, Costa A

EMDB-4693: 
Structure of a human nucleosome at 3.5 A resolution
Method: single particle / : Wilson MD, Nans A, Costa A

EMDB-4960: 
PFV intasome - nucleosome strand transfer complex
Method: single particle / : Renault L, Maskell DP, Wilson MD, Cherepanov P, Costa A

PDB-6r0c: 
Human-D02 Nucleosome Core Particle with biotin-streptavidin label
Method: single particle / : Pye VE, Wilson MD, Cherepanov P, Costa A

PDB-6rny: 
PFV intasome - nucleosome strand transfer complex
Method: single particle / : Pye VE, Renault L, Maskell DP, Cherepanov P, Costa A

EMDB-4958: 
Negative stain EM 3D reconstruction of the UvrA-UvrB-DNA complex.
Method: single particle / : Swuec P, Renault L, Costa A
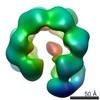
EMDB-3679: 
Full-length complex of S.cerevisiae Cdt1-MCM in apo state.
Method: single particle / : He J, Frigola J, Kinkelin K, Pye VE, Renault L, Douglas M, Remus D, Cherepanov P, Costa A, Diffley JFX
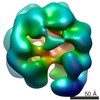
EMDB-3680: 
Full-length complex of S.cerevisiae Cdt1-MCM in ATPgS-bound state.
Method: single particle / : He J, Frigola J, Kinkelin K, Pye VE, Renault L, Douglas M, Remus D, Cherepanov P, Costa A, Diffley JFX
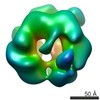
EMDB-3681: 
Full-length complex of S.cerevisiae MCM in ATPgS-bound state.
Method: single particle / : He J, Frigola J, Kinkelin K, Pye VE, Renault L, Douglas M, Remus D, Cherepanov P, Costa A, Diffley JFX

EMDB-3642: 
Full-length complex of CMG helicase with polymerase epsilon
Method: single particle / : Zhou JC, Janska A, Goswami P, Renault L, Abid Ali F, Kotecha A, Diffley JFX, Costa A

EMDB-3643: 
Complex of CMG helicase with polymerase epsilon lacking the catalytic domain of Pol2
Method: single particle / : Zhou JC, Janska A, Goswami P, Renault L, Abid Ali F, Kotecha A, Diffley JFX, Costa A
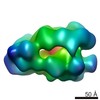
EMDB-3644: 
Full-length CMG helicase complex
Method: single particle / : Zhou JC, Janska A, Goswami P, Renault L, Abid Ali F, Kotecha A, Diffley JFX, Costa A

EMDB-3476: 
Structure of FANCI-FANCD2 hetero-dimer co-expressed with FANCC-FANCE-FANCF proteins.
Method: single particle / : Swuec P, Costa A

EMDB-3318: 
CryoEM structure of the CMG replicative helicase bound to a DNA fork (compact state)
Method: single particle / : Abid Ali F, Renault L, Costa A

EMDB-3319: 
CryoEM structure of the CMG replicative helicase bound to a DNA fork (relaxed state)
Method: single particle / : Abid Ali F, Renault L, Costa A

EMDB-3320: 
CryoEM structure of the ATP-treated CMG replicative helicase (compact state)
Method: single particle / : Renault L, Costa A

EMDB-3321: 
CryoEM structure of the ATP-treated CMG replicative helicase (relaxed state)
Method: single particle / : Renault L, Costa A

EMDB-2992: 
Structure of a pre-catalytic retroviral Intasome bound to a human nucleosome
Method: single particle / : Renault L, Maskell D, Cherepanov P, Costa A

EMDB-2772: 
CMG helicase bound to DNA and ATPgS
Method: single particle / : Costa A, Renault L, Swuec P, Petojevic T, Pesavento JJ, Ilves I, MacLellan-Gibson K, Fleck RA, Botchan MR, Berger JM
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
