+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Horse spleen apoferritin | |||||||||
 Map data Map data | horse spleen apo-ferritin | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | iron binding / iron release / iron regulation / METAL TRANSPORT | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.54 Å | |||||||||
 Authors Authors | Koning RI / Renault L | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Automated vitrification of cryo-EM samples with controllable sample thickness using suction and real-time optical inspection. Authors: Roman I Koning / Hildo Vader / Martijn van Nugteren / Peter A Grocutt / Wen Yang / Ludovic L R Renault / Abraham J Koster / Arnold C F Kamp / Michael Schwertner /   Abstract: The speed and efficiency of data collection and image processing in cryo-electron microscopy have increased over the last decade. However, cryo specimen preparation techniques have lagged and faster, ...The speed and efficiency of data collection and image processing in cryo-electron microscopy have increased over the last decade. However, cryo specimen preparation techniques have lagged and faster, more reproducible specimen preparation devices are needed. Here, we present a vitrification device with highly automated sample handling, requiring only limited user interaction. Moreover, the device allows inspection of thin films using light microscopy, since the excess liquid is removed through suction by tubes, not blotting paper. In combination with dew-point control, this enables thin film preparation in a controlled and reproducible manner. The advantage is that the quality of the prepared cryo specimen is characterized before electron microscopy data acquisition. The practicality and performance of the device are illustrated with experimental results obtained by vitrification of protein suspensions, lipid vesicles, bacterial and human cells, followed by imaged using single particle analysis, cryo-electron tomography, and cryo correlated light and electron microscopy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13738.map.gz emd_13738.map.gz | 12 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13738-v30.xml emd-13738-v30.xml emd-13738.xml emd-13738.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13738.png emd_13738.png | 140.6 KB | ||
| Filedesc metadata |  emd-13738.cif.gz emd-13738.cif.gz | 4.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13738 http://ftp.pdbj.org/pub/emdb/structures/EMD-13738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13738 | HTTPS FTP |
-Validation report
| Summary document |  emd_13738_validation.pdf.gz emd_13738_validation.pdf.gz | 369.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13738_full_validation.pdf.gz emd_13738_full_validation.pdf.gz | 369.3 KB | Display | |
| Data in XML |  emd_13738_validation.xml.gz emd_13738_validation.xml.gz | 6.3 KB | Display | |
| Data in CIF |  emd_13738_validation.cif.gz emd_13738_validation.cif.gz | 7.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13738 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13738 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13738 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13738 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13738.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13738.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | horse spleen apo-ferritin | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
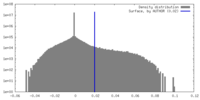
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : apoferritin
| Entire | Name: apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: apoferritin
| Supramolecule | Name: apoferritin / type: complex / ID: 1 / Parent: 0 / Details: Horse spleen apoferritin |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 504 MDa |
-Macromolecule #1: Apoferritin (equine spleen)
| Macromolecule | Name: Apoferritin (equine spleen) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: SSQIRQNYST EVEAAVNRLV NLYLRASYTY LSLGFYFDRD DVALEGVCHF FRELAEEKRE GAERLLKMQN QRGGRALFQD LQKPSQDEWG TTLDAMKAAI VLEKSLNQAL LDLHALGSAQ ADPHLCDFLE SHFLDEEVKL IKKMGDHLTN IQRLVGSQAG LGEYLFERLT LKHD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.018000000000000002 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 294 K / Instrument: HOMEMADE PLUNGER Details: Prototype of Linkam Plunger Sample applied by dipping grid in dispersion and excess solution removed by suction.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 3348 / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)