-Search query
-Search result
Showing 1 - 50 of 57 items for (author: puri & p)

EMDB-45105: 
Cryo-electron tomography of Candida glabrata plasma membrane
Method: electron tomography / : Jiang J, Keniya MV, Perlin DS, Dai W

EMDB-45106: 
Cryo-electron tomography of plasma membranes generated from caspofungin-treated Candida glabrata spheroplasts
Method: electron tomography / : Jiang J, Keniya MV, Perlin DS, Dai W

EMDB-45276: 
Map of full-length LPD-3 complex
Method: single particle / : Clark SA, Kang Y

EMDB-45399: 
Structure of the LPD-3 complex
Method: single particle / : Clark SA, Vanni S, Kang Y

EMDB-41024: 
MD65 N332-GT5 SOSIP in complex with RM_N332_03 Fab and RM20A3 Fab
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-41025: 
MD65 N332-GT5 SOSIP in complex with RM_N332_36 Fab and RM20A3 Fab
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-41026: 
MD65 N332-GT5 SOSIP in complex with RM_N332_32 Fab and RM20A3
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-41027: 
MD65 N332-GT5 SOSIP in complex with RM_N332_08 Fab and RM20A3 Fab
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-41034: 
MD64 N332-GT5 SOSIP
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-41035: 
MD65 N332-GT5 SOSIP in complex with RM_N332_07 Fab and RM20A3 Fab
Method: single particle / : Ozorowski G, Torres JL, Ward AB

PDB-8t49: 
MD65 N332-GT5 SOSIP in complex with RM_N332_03 Fab and RM20A3 Fab
Method: single particle / : Ozorowski G, Torres JL, Ward AB

PDB-8t4a: 
MD65 N332-GT5 SOSIP in complex with RM_N332_36 Fab and RM20A3 Fab
Method: single particle / : Ozorowski G, Torres JL, Ward AB

PDB-8t4b: 
MD65 N332-GT5 SOSIP in complex with RM_N332_32 Fab and RM20A3
Method: single particle / : Ozorowski G, Torres JL, Ward AB

PDB-8t4d: 
MD65 N332-GT5 SOSIP in complex with RM_N332_08 Fab and RM20A3 Fab
Method: single particle / : Ozorowski G, Torres JL, Ward AB

PDB-8t4k: 
MD64 N332-GT5 SOSIP
Method: single particle / : Ozorowski G, Torres JL, Ward AB

PDB-8t4l: 
MD65 N332-GT5 SOSIP in complex with RM_N332_07 Fab and RM20A3 Fab
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-16780: 
CryoEM structure of AL55 amyloid fibrils extracted from the kidney of an AL amyloidosis patient.
Method: helical / : Puri S, Schulte T, Chaves-Sanjuan A, Ricagno S

PDB-8cpe: 
CryoEM structure of AL55 amyloid fibrils extracted from the kidney of an AL amyloidosis patient.
Method: helical / : Puri S, Schulte T, Chaves-Sanjuan A, Ricagno S

EMDB-14004: 
Specific features and methylation sites of a plant 80S ribosome
Method: single particle / : Cottilli P, Itoh Y, Amunts A

PDB-7qiz: 
Specific features and methylation sites of a plant 80S ribosome
Method: single particle / : Cottilli P, Itoh Y, Amunts A

EMDB-14001: 
Specific features and methylation sites of a plant ribosome. 60S ribosomal subunit.
Method: single particle / : Cottilli P, Itoh Y, Amunts A

PDB-7qiw: 
Specific features and methylation sites of a plant ribosome. 60S ribosomal subunit.
Method: single particle / : Cottilli P, Itoh Y, Amunts A

EMDB-14002: 
Specific features and methylation sites of a plant ribosome. 40S body ribosomal subunit.
Method: single particle / : Cottilli P, Itoh Y, Amunts A

EMDB-14003: 
Specific features and methylation sites of a plant ribosome
Method: single particle / : Amunts A, Cottilli P

EMDB-14051: 
Specific features and methylation sites of a plant ribosome. 80S translating ribosome (A/A-P/P-E/E).
Method: single particle / : Cottilli P, Itoh Y, Amunts A

EMDB-14052: 
Specific features and methylation sites of a plant ribosome. 80S translating ribosome (A/P-P/E).
Method: single particle / : Cottilli P, Itoh Y, Amunts A

EMDB-32404: 
Specific features and methylation sites of a plant ribosome. 80S ribosome consensus map.
Method: single particle / : Cottilli P, Itoh Y, Amunts A

PDB-7qix: 
Specific features and methylation sites of a plant ribosome. 40S body ribosomal subunit.
Method: single particle / : Cottilli P, Itoh Y, Amunts A

PDB-7qiy: 
Specific features and methylation sites of a plant ribosome. 40S head ribosomal subunit.
Method: single particle / : Cottilli P, Itoh Y, Amunts A

EMDB-13326: 
T. maritima CorA in DDM micelles without Mg2+ bound in D2O
Method: single particle / : Larsen AH, Johansen NT, Arleth L

EMDB-13327: 
T. maritima CorA in DDM micelles with Mg2+ bound in D2O
Method: single particle / : Larsen AH, Johansen NT, Arleth L
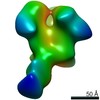
EMDB-22496: 
Base-directed response of rabbit #1 week 6 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB

EMDB-22498: 
N611-site directed response of rabbit #1 week 6 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB

EMDB-22499: 
Base-directed response of rabbit #1 week 12 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB
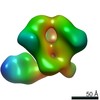
EMDB-22500: 
N611-site directed response of rabbit #1 week 12 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB
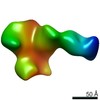
EMDB-22501: 
C3/V5-directed response of rabbit #1 week 12 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB
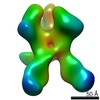
EMDB-22502: 
Base-directed response of rabbit #1 week 22 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB
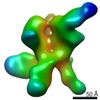
EMDB-22503: 
N611-site, base, and V1V3 directed response of rabbit #1 week 22 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB
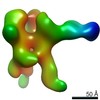
EMDB-22504: 
C3/V5 and base directed response of rabbit #1 week 22 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB

EMDB-22505: 
V1/V3 and base directed response of rabbit #1 week 22 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB
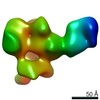
EMDB-23159: 
C3/V5-directed response of human donor G37080 polyclonal fragment antigen binding in complex with 1PGE-THIVC SOSIP
Method: single particle / : Sewell LM, Ozorowski G, Ward AB

EMDB-21009: 
Lon Protease from Yersinia pestis
Method: single particle / : Shin M, Puchades C

PDB-6v11: 
Lon Protease from Yersinia pestis
Method: single particle / : Shin M, Puchades C, Asmita A, Puri N, Adjei E, Wiseman RL, Karzai AW, Lander GC

EMDB-20133: 
Lon Protease from Yersinia pestis with Y2853 substrate
Method: single particle / : Shin M, Asmita A

PDB-6on2: 
Lon Protease from Yersinia pestis with Y2853 substrate
Method: single particle / : Shin M, Asmita A, Puchades C, Adjei E, Wiseman RL, Karzai AW, Lander GC

EMDB-4537: 
E. coli DnaBC apo complex
Method: single particle / : Arias-Palomo E, Puri N

EMDB-4538: 
E. coli DnaBC complex bound to ssDNA
Method: single particle / : Arias-Palomo E, Puri N

PDB-6qel: 
E. coli DnaBC apo complex
Method: single particle / : Arias-Palomo E, Puri N, O'Shea Murray VL, Yan Q, Berger JM

PDB-6qem: 
E. coli DnaBC complex bound to ssDNA
Method: single particle / : Arias-Palomo E, Puri N, O'Shea Murray VL, Yan Q, Berger JM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
