[English] 日本語
 Yorodumi
Yorodumi- EMDB-41025: MD65 N332-GT5 SOSIP in complex with RM_N332_36 Fab and RM20A3 Fab -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MD65 N332-GT5 SOSIP in complex with RM_N332_36 Fab and RM20A3 Fab | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | germline targeting / NHP / SOSIP / Env / HIV / BG18 / immunogen design / antibody / VIRAL PROTEIN / VIRAL PROTEIN-Immune System complex | |||||||||
| Biological species |    Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
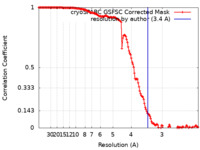
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Ozorowski G / Torres JL / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Vaccine priming of rare HIV broadly neutralizing antibody precursors in nonhuman primates. Authors: Jon M Steichen / Ivy Phung / Eugenia Salcedo / Gabriel Ozorowski / Jordan R Willis / Sabyasachi Baboo / Alessia Liguori / Christopher A Cottrell / Jonathan L Torres / Patrick J Madden / ...Authors: Jon M Steichen / Ivy Phung / Eugenia Salcedo / Gabriel Ozorowski / Jordan R Willis / Sabyasachi Baboo / Alessia Liguori / Christopher A Cottrell / Jonathan L Torres / Patrick J Madden / Krystal M Ma / Henry J Sutton / Jeong Hyun Lee / Oleksandr Kalyuzhniy / Joel D Allen / Oscar L Rodriguez / Yumiko Adachi / Tina-Marie Mullen / Erik Georgeson / Michael Kubitz / Alison Burns / Shawn Barman / Rohini Mopuri / Amanda Metz / Tasha K Altheide / Jolene K Diedrich / Swati Saha / Kaitlyn Shields / Steven E Schultze / Melissa L Smith / Torben Schiffner / Dennis R Burton / Corey T Watson / Steven E Bosinger / Max Crispin / John R Yates / James C Paulson / Andrew B Ward / Devin Sok / Shane Crotty / William R Schief /   Abstract: Germline-targeting immunogens hold promise for initiating the induction of broadly neutralizing antibodies (bnAbs) to HIV and other pathogens. However, antibody-antigen recognition is typically ...Germline-targeting immunogens hold promise for initiating the induction of broadly neutralizing antibodies (bnAbs) to HIV and other pathogens. However, antibody-antigen recognition is typically dominated by heavy chain complementarity determining region 3 (HCDR3) interactions, and vaccine priming of HCDR3-dominant bnAbs by germline-targeting immunogens has not been demonstrated in humans or outbred animals. In this work, immunization with N332-GT5, an HIV envelope trimer designed to target precursors of the HCDR3-dominant bnAb BG18, primed bnAb-precursor B cells in eight of eight rhesus macaques to substantial frequencies and with diverse lineages in germinal center and memory B cells. We confirmed bnAb-mimicking, HCDR3-dominant, trimer-binding interactions with cryo-electron microscopy. Our results demonstrate proof of principle for HCDR3-dominant bnAb-precursor priming in outbred animals and suggest that N332-GT5 holds promise for the induction of similar responses in humans. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41025.map.gz emd_41025.map.gz | 259.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41025-v30.xml emd-41025-v30.xml emd-41025.xml emd-41025.xml | 27.2 KB 27.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41025_fsc.xml emd_41025_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_41025.png emd_41025.png | 110.2 KB | ||
| Masks |  emd_41025_msk_1.map emd_41025_msk_1.map | 274.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41025.cif.gz emd-41025.cif.gz | 8 KB | ||
| Others |  emd_41025_half_map_1.map.gz emd_41025_half_map_1.map.gz emd_41025_half_map_2.map.gz emd_41025_half_map_2.map.gz | 255.1 MB 255.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41025 http://ftp.pdbj.org/pub/emdb/structures/EMD-41025 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41025 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41025 | HTTPS FTP |
-Related structure data
| Related structure data |  8t4aMC  8t49C  8t4bC  8t4dC  8t4kC  8t4lC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41025.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41025.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41025_msk_1.map emd_41025_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_41025_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_41025_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MD65 N332-GT5 SOSIP in complex with RM_N332_36 Fab and RM20A3 Fab
| Entire | Name: MD65 N332-GT5 SOSIP in complex with RM_N332_36 Fab and RM20A3 Fab |
|---|---|
| Components |
|
-Supramolecule #1: MD65 N332-GT5 SOSIP in complex with RM_N332_36 Fab and RM20A3 Fab
| Supramolecule | Name: MD65 N332-GT5 SOSIP in complex with RM_N332_36 Fab and RM20A3 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: MD65 N332-GT5 SOSIP gp120
| Macromolecule | Name: MD65 N332-GT5 SOSIP gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.286773 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHEDIIS LWDQSLKPC VKLTPLCVTL QCTNYAPKLR SMMRGEIKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHEDIIS LWDQSLKPC VKLTPLCVTL QCTNYAPKLR SMMRGEIKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SAITQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CQNVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV IIRSENI TNNAKNILVQ LNTSVQINCT RPSNNTVKSI RIGPGQAFYY FGDVLGHVRM AHCNISKATW NETLGKVVKQ LRK HFGNNT IIRFAQSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSLILPC WIKQIINMWQ RIGQ AMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRTVG RRRRR R |
-Macromolecule #2: RM20A3 heavy chain Fv
| Macromolecule | Name: RM20A3 heavy chain Fv / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.511111 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVETGGG LVQPGGSLKL SCRASGYTFS SFAMSWVRQA PGKGLEWVSL INDRGGLTFY VDSVKGRFTI SRDNSKNTLS LQMHSLRDG DTAVYYCATG GMSSALQSSK YYFDFWGQGA LVTVSS |
-Macromolecule #3: RM20A3 light chain Fv
| Macromolecule | Name: RM20A3 light chain Fv / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.5088 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ALTQPPSVSG SPGQSVTISC TGTSSDIGSY NYVSWYQQHP GKAPKLMIYD VTQRPSGVSD RFSGSKSGNT ASLTISGLQA DDEADYYCS AYAGRQTFYI FGGGTRLTVL GQPKASPTVT LFPPSSEEL |
-Macromolecule #4: MD65 N332-GT5 SOSIP gp41
| Macromolecule | Name: MD65 N332-GT5 SOSIP gp41 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.096148 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AAGIGASSDG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEP QQHLLKDTHW GIKQLQARVL AVEHYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD |
-Macromolecule #5: RM_N332_36 heavy chain Fv
| Macromolecule | Name: RM_N332_36 heavy chain Fv / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.779726 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVTLKESGPA LVKPTQTLTL TCTFSGFSLS TNYMGVDWIR QPPGKALEWL ARIYWNDDKY YSTSLKSRLT ISKDTSKNQV VLTMSNMDP VDTATYYCAR VKLSVWGVLQ FFEWSDYGLD SWGQGVVVTV SS |
-Macromolecule #6: RM_N332_36 light chain Fv
| Macromolecule | Name: RM_N332_36 light chain Fv / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.40742 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SYVLTQPPSV SVSPGQTARI TCGADNIGSQ SVHWYQQKSP QAPVLVISAD TERPSGIPER FSGSNSGNTA TLTISGVEAG DEADYYCQV WDSDSKYALF GGGTRLTVL |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 51 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)