+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20133 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lon Protease from Yersinia pestis with Y2853 substrate | ||||||||||||||||||
 Map data Map data | Composite stitched map for final model building and refinement | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Lon / mitochondrial protease / AAA+ / ATPase / HYDROLASE | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationendopeptidase La / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / cellular response to heat / sequence-specific DNA binding / serine-type endopeptidase activity / ATP hydrolysis activity / ATP binding / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||||||||
 Authors Authors | Shin M / Asmita A | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2020 Journal: Sci Adv / Year: 2020Title: Structural basis for distinct operational modes and protease activation in AAA+ protease Lon. Authors: Mia Shin / Cristina Puchades / Ananya Asmita / Neha Puri / Eric Adjei / R Luke Wiseman / A Wali Karzai / Gabriel C Lander /  Abstract: Substrate-bound structures of AAA+ protein translocases reveal a conserved asymmetric spiral staircase architecture wherein a sequential ATP hydrolysis cycle drives hand-over-hand substrate ...Substrate-bound structures of AAA+ protein translocases reveal a conserved asymmetric spiral staircase architecture wherein a sequential ATP hydrolysis cycle drives hand-over-hand substrate translocation. However, this configuration is unlikely to represent the full conformational landscape of these enzymes, as biochemical studies suggest distinct conformational states depending on the presence or absence of substrate. Here, we used cryo-electron microscopy to determine structures of the Lon AAA+ protease in the absence and presence of substrate, uncovering the mechanistic basis for two distinct operational modes. In the absence of substrate, Lon adopts a left-handed, "open" spiral organization with autoinhibited proteolytic active sites. Upon the addition of substrate, Lon undergoes a reorganization to assemble an enzymatically active, right-handed "closed" conformer with active protease sites. These findings define the mechanistic principles underlying the operational plasticity required for processing diverse protein substrates. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20133.map.gz emd_20133.map.gz | 3.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20133-v30.xml emd-20133-v30.xml emd-20133.xml emd-20133.xml | 38.4 KB 38.4 KB | Display Display |  EMDB header EMDB header |
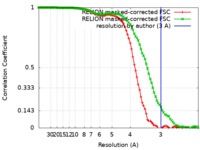
| FSC (resolution estimation) |  emd_20133_fsc_1.xml emd_20133_fsc_1.xml emd_20133_fsc_2.xml emd_20133_fsc_2.xml | 8.6 KB 8.6 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_20133.png emd_20133.png | 68 KB | ||
| Filedesc metadata |  emd-20133.cif.gz emd-20133.cif.gz | 8.2 KB | ||
| Others |  emd_20133_additional_1.map.gz emd_20133_additional_1.map.gz emd_20133_additional_2.map.gz emd_20133_additional_2.map.gz emd_20133_additional_3.map.gz emd_20133_additional_3.map.gz emd_20133_additional_4.map.gz emd_20133_additional_4.map.gz emd_20133_half_map_1.map.gz emd_20133_half_map_1.map.gz emd_20133_half_map_2.map.gz emd_20133_half_map_2.map.gz | 7.8 MB 7.8 MB 7.8 MB 7.8 MB 7.8 MB 7.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20133 http://ftp.pdbj.org/pub/emdb/structures/EMD-20133 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20133 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20133 | HTTPS FTP |
-Related structure data
| Related structure data |  6on2MC  6v11C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20133.map.gz / Format: CCP4 / Size: 8.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20133.map.gz / Format: CCP4 / Size: 8.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite stitched map for final model building and refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Half-map of E and F subunits used to...
| File | emd_20133_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of E and F subunits used to make composite stitched map for final model building and refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
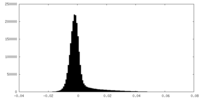
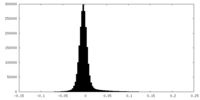
| Density Histograms |
-Additional map: Half-map of E and F subunits used to...
| File | emd_20133_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of E and F subunits used to make composite stitched map for final model building and refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
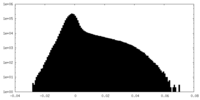
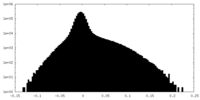
| Density Histograms |
-Additional map: Filtered map of E and F subunits used...
| File | emd_20133_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Filtered map of E and F subunits used to make composite stitched map for final model building and refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
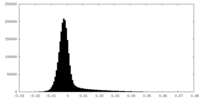
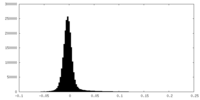
| Density Histograms |
-Additional map: Filtered map of A, B, C, and D...
| File | emd_20133_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Filtered map of A, B, C, and D subunits used to make composite stitched map for final model building and refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
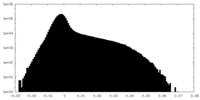
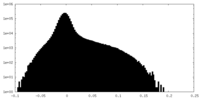
| Density Histograms |
-Half map: Half-map of A, B, C, and D subunits...
| File | emd_20133_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of A, B, C, and D subunits used to make composite stitched map for final model building and refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map of A, B, C, and D subunits...
| File | emd_20133_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of A, B, C, and D subunits used to make composite stitched map for final model building and refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Lon protease bound to Y2853 substrate
| Entire | Name: Lon protease bound to Y2853 substrate |
|---|---|
| Components |
|
-Supramolecule #1: Lon protease bound to Y2853 substrate
| Supramolecule | Name: Lon protease bound to Y2853 substrate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Complexes consisting of homohexameric Lon protease from Yersinia pestis bound to Y2853 substrate were isolated using size-exclusion chromatography |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP-dependent protease La
| Macromolecule | Name: ATP-dependent protease La / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: endopeptidase La |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.927051 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ALKRKIEAAK MPKDAREKTE AELQKLKMMS PMSAEATVVR GYIDWMLQVP WNSRSKVKKD LVKAQEVLDT DHYGLERVKD RILEYLAVQ SRVSKIKGPI LCLVGPPGVG KTSLGQSIAK ATGRQYVRMA LGGVRDEAEI RGHRRTYIGS MPGKLIQKMA K VGVKNPLF ...String: ALKRKIEAAK MPKDAREKTE AELQKLKMMS PMSAEATVVR GYIDWMLQVP WNSRSKVKKD LVKAQEVLDT DHYGLERVKD RILEYLAVQ SRVSKIKGPI LCLVGPPGVG KTSLGQSIAK ATGRQYVRMA LGGVRDEAEI RGHRRTYIGS MPGKLIQKMA K VGVKNPLF LLDQIDKMAS DMRGDPASAL LEVLDPEQNV AFNDHYLEVD YDLSDVMFVA TSNSMNIPAP LLDRMEVIRL SG YTEDEKL NIAKQHLLPK QFERNAIKKG ELTIDDSAIM SIIRYYTREA GVRSLEREIS KLCRKAVKNL LMDKTVKHIE ING DNLKDF LGVQKVDYGR ADTENRVGQV TGLAWTEVGG DLLTIETACV PGKGKLTYTG SLGEVMQESI QAALTVVRAR ADKL GINPD FYEKRDIHVH VPEGATPKDG PSAGIAMCTA LVSCLTGNPV RADVAMTGEI TLRGLVLPIG GLKEKLLAAH RGGIK VVLI PDDNKRDLEE IPDNVIADLE IHPVKRIDDV LAIALEHPAF UniProtKB: Lon protease |
-Macromolecule #2: Bound Y2853 Substrate
| Macromolecule | Name: Bound Y2853 Substrate / type: protein_or_peptide / ID: 2 Details: Y2853 substrate was added to Lon and modeled here as a polyalanine chain Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 515.56 Da |
| Recombinant expression | Organism:  |
| Sequence | String: AAAAAAA |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.95 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: Solutions were made fresh from concentrated and filtered using a 0.1 um syringe filter to avoid microbial contamination. Buffers were stored on ice and used within 15 minutes of mixing in ...Details: Solutions were made fresh from concentrated and filtered using a 0.1 um syringe filter to avoid microbial contamination. Buffers were stored on ice and used within 15 minutes of mixing in order to avoid excess ATP hydrolysis. | ||||||||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.01 kPa Details: Grids were plasma treated for 30 seconds using a 15 mA current operating under atmospheric gases using a glow discharger (Electron Microscopy Sciences). | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER Details: 4 uL of sample was applied per grid and manually blotted for 4 seconds followed by immediately plunge-freezing in liquid ethane cooled by liquid nitrogen.. | ||||||||||||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 80.0 K / Max: 90.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.14 mrad |
| Details | Coma-free alignment procedure from Herzik & Wu, Nature Methods (2017). Preliminary grid screening was performed manually prior to data collection. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 0-43 / Number grids imaged: 2 / Number real images: 4071 / Average exposure time: 11.0 sec. / Average electron dose: 52.0 e/Å2 Details: Images were collected in counting mode at 4 frames per second |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 1.5 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 43478 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Initial homology model was built using SWISS-MODEL and initial rigid body docking was done using UCSF Chimera |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 52 / Target criteria: Correlation coefficient |
| Output model |  PDB-6on2: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)