+Search query
-Structure paper
| Title | Elicitation of potent serum neutralizing antibody responses in rabbits by immunization with an HIV-1 clade C trimeric Env derived from an Indian elite neutralizer. |
|---|---|
| Journal, issue, pages | PLoS Pathog, Vol. 17, Issue 4, Page e1008977, Year 2021 |
| Publish date | Apr 7, 2021 |
 Authors Authors | Rajesh Kumar / Suprit Deshpande / Leigh M Sewall / Gabriel Ozorowski / Christopher A Cottrell / Wen-Hsin Lee / Lauren G Holden / Sara T Richey / Antra Singh Chandrawacar / Kanika Dhiman / Ashish / Vivek Kumar / Shubbir Ahmed / Nitin Hingankar / Naresh Kumar / Kailapuri G Murugavel / Aylur K Srikrishnan / Devin Sok / Andrew B Ward / Jayanta Bhattacharya /   |
| PubMed Abstract | Evaluating the structure-function relationship of viral envelope (Env) evolution and the development of broadly cross-neutralizing antibodies (bnAbs) in natural infection can inform rational ...Evaluating the structure-function relationship of viral envelope (Env) evolution and the development of broadly cross-neutralizing antibodies (bnAbs) in natural infection can inform rational immunogen design. In the present study, we examined the magnitude and specificity of autologous neutralizing antibodies induced in rabbits by a novel HIV-1 clade C Env protein (1PGE-THIVC) vis-à-vis those developed in an elite neutralizer from whom the env sequence was obtained that was used to prepare the soluble Env protein. The novel 1PGE-THIVC Env trimer displayed a native like pre-fusion closed conformation in solution as determined by small angle X-ray scattering (SAXS) and negative stain electron microscopy (EM). This closed spike conformation of 1PGE-THIVC Env trimers was correlated with weak or undetectable binding of non-neutralizing monoclonal antibodies (mAbs) compared to neutralizing mAbs. Furthermore, 1PGE-THIVC SOSIP induced potent neutralizing antibodies in rabbits to autologous virus variants. The autologous neutralizing antibody specificity induced in rabbits by 1PGE-THIVC was mapped to the C3/V4 region (T362/P401) of viral Env. This observation agreed with electron microscopy polyclonal epitope mapping (EMPEM) of the Env trimer complexed with IgG Fab prepared from the immunized rabbit sera. Our study demonstrated neutralization of sequence matched and unmatched autologous viruses by serum antibodies induced in rabbits by 1PGE-THIVC and also highlighted a comparable specificity for the 1PGE-THIVC SOSIP trimer with that seen with polyclonal antibodies elicited in the elite neutralizer by negative-stain electron microscopy polyclonal epitope (ns-EMPEM) mapping. |
 External links External links |  PLoS Pathog / PLoS Pathog /  PubMed:33826683 / PubMed:33826683 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 25.0 - 29.0 Å |
| Structure data | 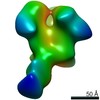 EMDB-22496:  EMDB-22498:  EMDB-22499: 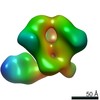 EMDB-22500: 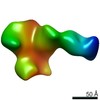 EMDB-22501: 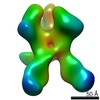 EMDB-22502:  EMDB-22503:  EMDB-22504:  EMDB-22505:  EMDB-23159: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




 Homo sapiens (human)
Homo sapiens (human)