-Search query
-Search result
Showing all 33 items for (author: nicholson & d)

EMDB-47975: 
Cryo-EM structure of the portal-tail complex of LME-1 phage
Method: single particle / : Deme JC, Lea SM

EMDB-47984: 
Cryo-EM structure of the icosahedral capsid of LME-1 phage
Method: single particle / : Deme JC, Lea SM

EMDB-29281: 
Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

EMDB-29282: 
Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

PDB-8flk: 
Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

PDB-8flm: 
Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

EMDB-16540: 
Neurofascin isoform NF155 extracellular domain
Method: single particle / : McKie SJ, Deane JE, Butt BG

EMDB-13936: 
Cryo-EM structure of the ribosome from Encephalitozoon cuniculi
Method: single particle / : Nicholson D, Ranson NA

PDB-7qep: 
Cryo-EM structure of the ribosome from Encephalitozoon cuniculi
Method: single particle / : Nicholson D, Ranson NA, Melnikov SV

EMDB-13191: 
Staphylococcus aureus ribosome in complex with Sal(B)
Method: single particle / : Nicholson D, Ranson NA

PDB-7p48: 
Staphylococcus aureus ribosome in complex with Sal(B)
Method: single particle / : Nicholson D, Ranson NA, O'Neill AJ

EMDB-10809: 
Acinetobacter baumannii ribosome-amikacin complex - 50S subunit
Method: single particle / : Nicholson D, Edwards TA, O'Neill AJ, Ranson NA

EMDB-10869: 
Acinetobacter baumannii ribosome-amikacin complex - 30S subunit body
Method: single particle / : Nicholson D, Edwards TA, O'Neill AJ, Ranson NA

EMDB-10892: 
Acinetobacter baumannii ribosome-amikacin complex - 30S subunit head
Method: single particle / : Nicholson D, Edwards TA

EMDB-10898: 
Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit
Method: single particle / : Nicholson D, Edwards TA

EMDB-10914: 
Acinetobacter baumannii ribosome-tigecycline complex - 30S subunit body
Method: single particle / : Nicholson D, Edwards TA, O'Neill AJ, Ranson NA

EMDB-10915: 
Acinetobacter baumannii ribosome-tigecycline complex - 30S subunit head
Method: single particle / : Nicholson D, Edwards TA, O'Neill AJ, Ranson NA

PDB-6yhs: 
Acinetobacter baumannii ribosome-amikacin complex - 50S subunit
Method: single particle / : Nicholson D, Edwards TA, O'Neill AJ, Ranson NA

PDB-6ypu: 
Acinetobacter baumannii ribosome-amikacin complex - 30S subunit body
Method: single particle / : Nicholson D, Edwards TA, O'Neill AJ, Ranson NA

PDB-6ys5: 
Acinetobacter baumannii ribosome-amikacin complex - 30S subunit head
Method: single particle / : Nicholson D, Edwards TA, O'Neill AJ, Ranson NA

PDB-6ysi: 
Acinetobacter baumannii ribosome-tigecycline complex - 50S subunit
Method: single particle / : Nicholson D, Edwards TA, O'Neill AJ, Ranson NA

PDB-6yt9: 
Acinetobacter baumannii ribosome-tigecycline complex - 30S subunit body
Method: single particle / : Nicholson D, Edwards TA, O'Neill AJ, Ranson NA

PDB-6ytf: 
Acinetobacter baumannii ribosome-tigecycline complex - 30S subunit head
Method: single particle / : Nicholson D, Edwards TA, O'Neill AJ, Ranson NA

EMDB-8141: 
Cryo-EM structure of the human FANCD2/FANCI complex reveals a novel Tower domain required for FANCD2 monoubiquitination- Full length FANCD2/FANCI
Method: single particle / : Liang CC, Li Z, Nicholson WV, Venien-Bryan C, Cohn MA

EMDB-8142: 
Cryo-EM structure of the human FANCD2/FANCI complex reveals a novel Tower domain required for FANCD2 monoubiquitination- FANCD2-minusTower/FANCI complex
Method: single particle / : Liang CC, Li Z, Nicholson WV, Venien-Bryan C, Cohn MA
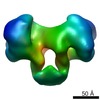
EMDB-1890: 
EcoR124 Type I DNA restriction-modification enzyme complex in closed state with bound 30bp cognate DNA fragment. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

EMDB-1891: 
EcoR124 Type I DNA restriction-modification enzyme complex without DNA (open state). Low resolution 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF
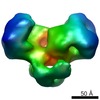
EMDB-1892: 
EcoR124 Type I DNA restriction-modification enzyme complex (in closed state) with bound DNA mimic protein Ocr from phage T7. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

EMDB-1893: 
EcoKI Type I DNA restriction-modification enzyme complex in closed state with bound 75bp cognate DNA fragment. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

EMDB-1748: 
Asymmetric organization of connexin26 gap junction channels
Method: electron crystallography / : Oshima A, Tani K, Toloue MM, Hiroaki Y, Smock A, Inukai S, Cone A, Nicholson BJ, Sosinsky GE, Fujiyoshi Y

EMDB-1749: 
A structure of Cx26M34Adel2-7 at 10 angstrom resolution
Method: electron crystallography / : Oshima A, Tani K, Toloue MM, Hiroaki Y, Smock A, Inukai S, Cone A, Nicholson BJ, Sosinsky GE, Fujiyoshi Y

PDB-3iz1: 
C-alpha model fitted into the EM structure of Cx26M34A
Method: electron crystallography / : Oshima A, Tani K, Toloue MM, Hiroaki Y, Smock A, Inukai S, Cone A, Nicholson BJ, Sosinsky GE, Fujiyoshi Y

PDB-3iz2: 
C-alpha model fitted into the EM structure of Cx26M34Adel2-7
Method: electron crystallography / : Oshima A, Tani K, Toloue MM, Hiroaki Y, Smock A, Inukai S, Cone A, Nicholson BJ, Sosinsky GE, Fujiyoshi Y
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
